I. What are the Enzymes and Processes in RNA Transcription?
Hopefully after covering the last section, you now have a better understanding and the fundamentals before diving into the actual enzymes and processes involved in RNA transcription – you’ve got the building blocks, now let’s get into the nitty gritty!
Undeniably, RNA transcription has its own facets that makes learning it both a little easier and harder when compared to DNA replication. While we still deal with DNA for a portion of this process, we get into more complex topics such as splicing and post-transcriptional modifications.
Luckily, as a little spoiler, RNA transcription contains MUCH less enzymes than DNA replication! Actually, much of the topics and items you should know about actually takes place post transcription, which will talk about more going further in the article!
II. Mechanism of Transcription
As stated above, there are much less enzymes to memorize for RNA transcription; in fact, there’s only 1: RNA polymerase! Look to the figure bel
A. Transcriptional Enzymes and Processes
As depicted below, transcription is actually relatively not as complicated as DNA replication, being that there’s only 1 enzyme to worry about! Let’s take a look at RNA polymerase in a more detailed sense.
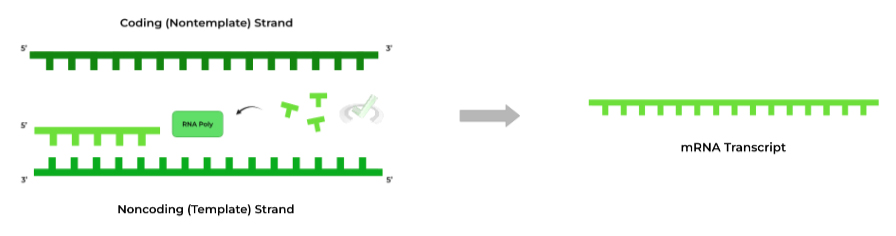
I. RNA Polymerase
Similar to DNA polymerase, RNA polymerase also catalyzes the addition of nucleotides through a similar nucleophilic substitution mechanism; however, now it’s attaching ribonucleotides instead of deoxyribonucleotides.
In addition, there are other aspects that make RNA polymerase a bit more unique! RNA polymerase has actually evolved to have additional helicase function, so there is no need for DNA helicase in transcription!
Furthermore, unlike DNA polymerase, RNA polymerase DOES NOT require a primer in order to initiate RNA transcription! Look at a table of a concise summary!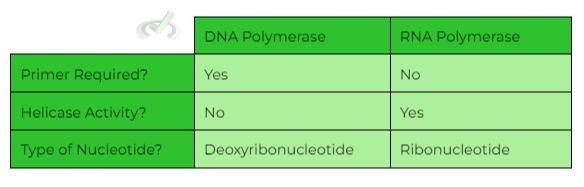
Also note that specifically, RNA polymerase II is utilized for mRNA transcription! There are other types like RNA polymerase I and III, but you’ll most likely be tested on type II as mentioned above.
Other important proteins involved in transcription are transcription factors, which will be covered in more detail in our “Gene Expression: Eukaryotes” article!
B. Post Transcriptional Modifications: mRNA Processing in Eukaryotes
Funny enough, the complex transcription topics actually take place after transcription! Before ribosomal translation, mRNAmust first undergo modifications in order to become mature mRNA.
Prior to post-transcriptional modification and maturation of mRNA, we use the term pre-mRNA for the mRNA that is not yet matured.
There are 3 main types of pre-mRNA processing modifications: 5’-methylguanosine cap, 3’-polyA-tail addition, and alternative splicing!
I. 5’-methylguanosine cap & 3’-polyA-tail addition
We’ve decided to combine these 2 modifications together because their functions are very similar.
The 5’-methylguanosine cap is composed of a guanosine nucleoside with a methyl group attached and the 3’-polyA-tail is a long sequence of adenine nucleotides at the 3’ end of the mRNA.

These additions to the 5’ and 3’ end protect the mRNA transcript from degradation in the cytosol by possible exonucleases. In addition, the 5’ cap also acts as a ribosomal recognition site to begin translation.
II. Alternative Splicing: Spliceosome and Ribozymes
Before getting into the nuances of alternative splicing, let’s get a more detailed explanation about genes and pre-mRNA structure.
Within genes, there are certain sequences which DO NOT code for any protein structure and sequences which DO code for protein structure. These sequences are termed introns and exons, respectively.
Because RNA polymerase cannot differentiate between introns & exons, both of these sequences carry on into the mRNA during transcription.
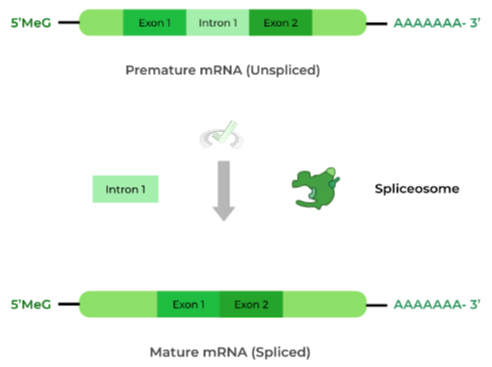
Before the mRNA is translated, the introns must be “spliced out” so that only the exons remain!
The spliceosome is a protein-ribozyme complex that is responsible for splicing out the introns, as shown above. This makes sense as we only want to include sequences that are coding for proteins (i.e. exons)!
III. Bridge/Overlap
Because of alternative splicing, the exons can be spliced in many different variations! From these variations can come types of mRNAs and protein isoforms! Let’s see how this may happen!
I. Variation in Protein Isoform via Alternative Splicing
Recall that the spliceosome primarily works to splice out introns in the pre-mRNA. Additionally, the spliceosome can also rearrange the orders of the exons, which results in the variation in protein isoforms!
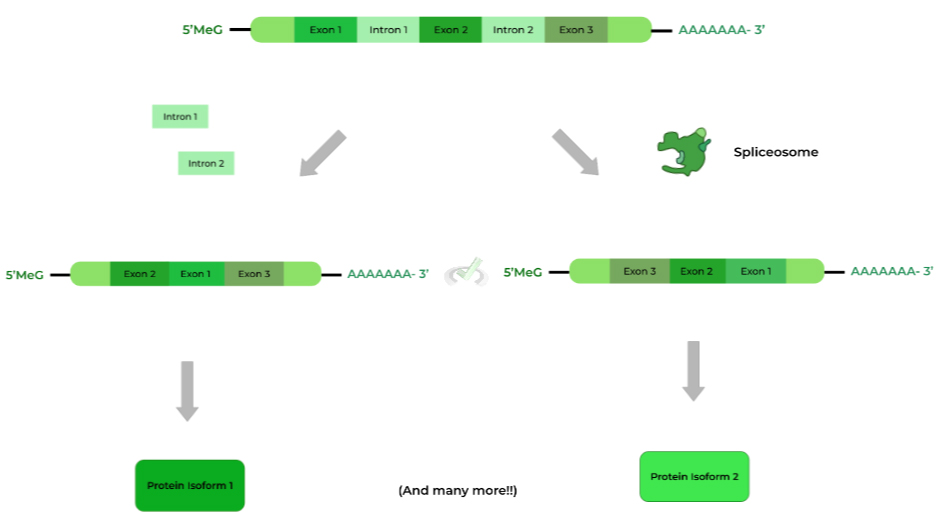
One real world example of how alternative splicing can be applied is in GLUT proteins, which are involved in the transport of glucose into the cell and come in 4 different protein isoforms: GLUT 1, 2, 3, and 4.
With the different isoforms also come slight differences in their characteristics: For example, GLUT 2 is primarily localized within the liver while GLUT 4 is found in abundance in the skeletal muscle!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Transcriptional Enzymes and Processes
RNA transcription involves the synthesis of mRNA transcripts from genetic DNA sequences which code for proteins, RNA structures, etc.
I. RNA Polymerase
The main enzyme involved in RNA transcription is RNA polymerase II, which has both helicase and polymerase activity to unwind and transcribe the DNA!
Note also that unlike DNA polymerase, RNA polymerase DOES NOT require a primer and can add nucleotides without the need of an existing strand!B. Post Transcriptional Modifications: mRNA Processing in Eukaryotes
After transcription, the mRNA (termed “pre-mRNA” at this stage) still needs to undergo various modifications including the addition of the 5’-methylguanosine cap & the 3’-polyA-tail as well as alternative splicing.
I. 5’-methylguanosine cap & 3’-polyA-tail addition
These additions to the mRNA transcript primarily protect the mRNA from degradation from exonucleases in the cytoplasm. In addition, the 5’-methylguanosine cap serves as a ribosomal recognition site to begin translation!
II. Alternative Splicing: Spliceosome and Ribozymes
Because pre-mRNA still contains introns (noncoding sequences), the spliceosome, a protein-ribozyme complex, is responsible for “splicing” out the introns so that the mature mRNA only contains exons!
In addition, alternative splicing also allows for the generation of various isoforms of proteins because the exons can be spliced together in many different variations!V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
In what ways are DNA replication and RNA transcription similar given the following choices below?
I. Use of template strand
II. DNA Helicase Involvement
III. Polymerase Involvement
A. I only
B. II only
C. II and III
D. I and III
Ans. D
Both DNA replication and RNA transcription will both use template strands in order to generate daughter DNA and mRNA respectively. Actually, DNA replication has to use 2 template strands because it needs to generate the double stranded DNA helix.
Likewise, polymerase involvement is found in both processes in the form of DNA and RNA polymerase.
However, DNA helicase involvement only takes place in DNA replication as RNA polymerase has embedded helicase function and doesn’t require the presence of DNA helicase for transcription.
Sample Practice Question 2
Suppose there is a defective mutation in the spliceosome which renders it nonfunctional. Given this, how would the resulting mRNA now compare in length to the normally spliced mRNA?
A. It would be smaller.
B. It would be bigger.
C. It would be the same.
D. Not enough information to determine.
Ans. B
Recall that the spliceosome is responsible for splicing out the introns from the mRNA transcript in order for it to be translated by the ribosome. As a result, because the introns are taken out, mature mRNA is smaller than the pre-mRNA.
However, with a nonfunctional mutation in the spliceosome, the introns now cannot be spliced out and will remain in the mRNA. Thus, the mRNA is much larger than the spliced mRNA because it now has additional introns contributing to the length.



 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these