I. What are the Enzymes and Processes in DNA Replication?
We are so sorry as this article contains just a portion of the many enzymes that need to be memorized for the MCAT. 
You’ve maybe heard of these enzymes before if you took an introductory biology course whether in college or even in high school such as DNA polymerase, helicase, etc. Nevertheless, we’ll for sure give you all an overview with all the necessary information while keeping it succinct.
We really want to stress relating this article and the enzymes covered back to the previous “DNA Replication: Key Terms” article, as that article really provides the foundation for how to understand the function of the enzymes and the role in DNA replication!
II. DNA Replication Overview
We’ll divide the content review section in 2 ways: 1) a grand, general overview of the enzymes involved in DNA replication & 2) a specific breakdown of the more high yield DNA replication enzymes.
A. General Mechanism of Replication Overview
Here’s a general diagram of DNA replication containing all the relevant enzymes involved in the replication process!
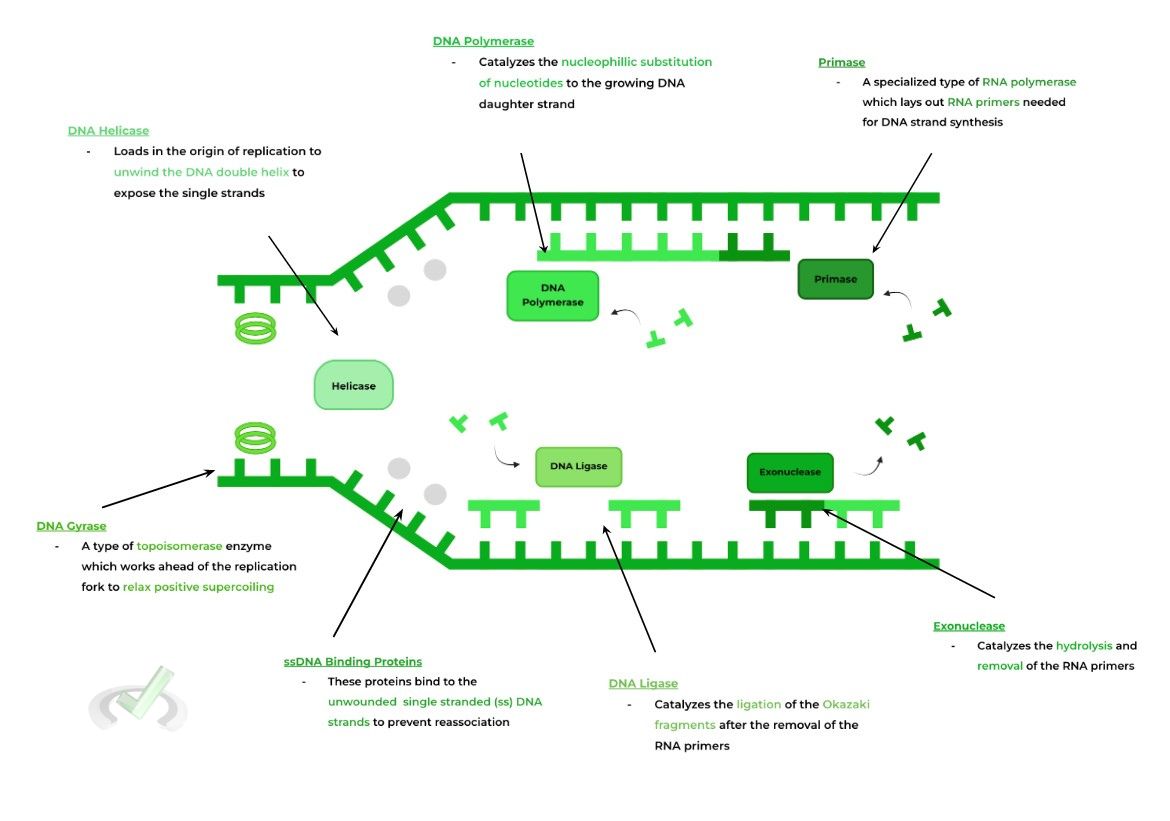
B. Specific Enzymes Involved in Replication
Let’s move on to highlighting 4 of the more high yield enzymes involved in DNA replication! As listed below, we’ll focus on: Primase, DNA Polymerase, Exonuclease, and DNA Ligase.
I. Primase
As mentioned above, primase is actually a type of RNA polymerase, which is funny as we’re dealing with DNA replication! Though a little confusing at first, let’s try to break this all down!
Interestingly enough, RNA primers are needed to initiate DNA synthesis because DNA polymerases have not evolved to initiate synthesis instantly without the need of a precursor.
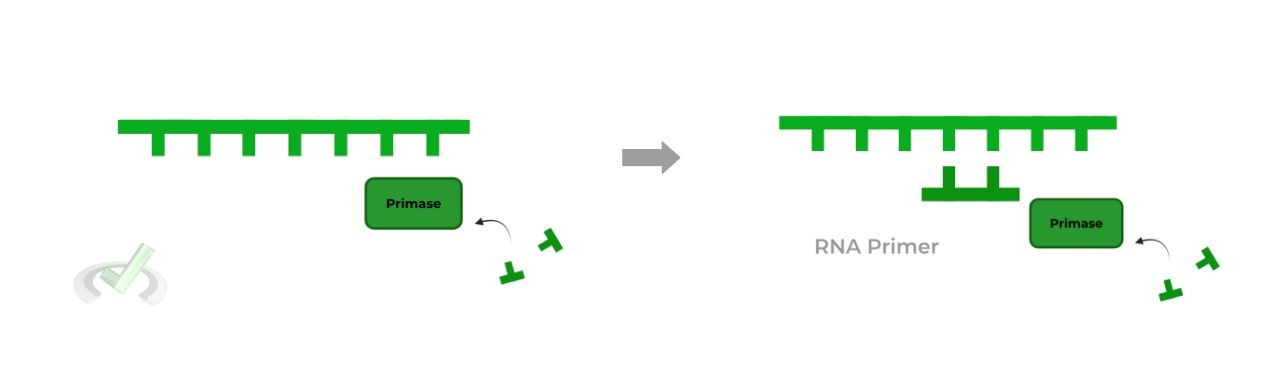
In addition, as of now it’s up to debate and research to why DNA primers are not used, where some research has cited that eukaryotic cells simply don’t have DNA primers. For now, just commit to memory that RNA primers are required.
II. DNA Polymerase III
Though all the enzymes are important for DNA replication, this enzyme is without a doubt probably the most important (though we’re not trying to pick favorites!!).
This is due to its function, as it catalyzes the attachment of incoming nucleotides to the growing daughter stand! The specific reaction is actually a nucleophilic substitution reaction!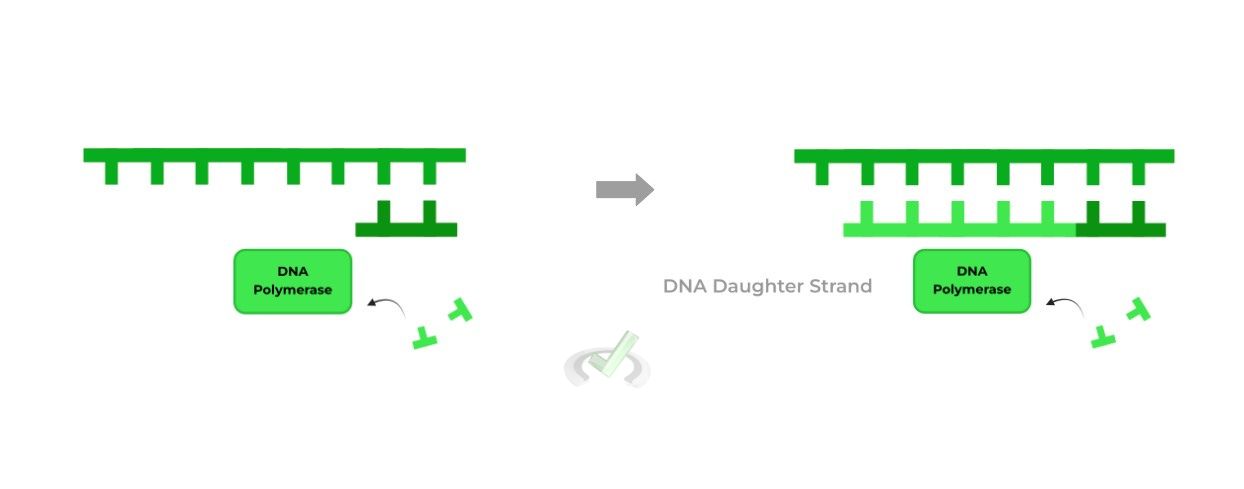
One high yield fact to understand about DNA polymerase is that it only reads template strands IN THE 3’-5’ DIRECTION. This is why, as stated before, daughter DNA strands are synthesized in the 5’-3’ direction.
The type of DNA polymerase used also differs between eukaryotes and prokaryotes! Eukaryotes will actually use a combination consisting of DNA polymerase 𝛼, 𝛿, and 𝟄 while prokaryotes use DNA polymerase III.III. Exonuclease
It’s crucial that the RNA primers are removed during the process after they’ve been used as a starting point to ensure only DNA is present! These exonuclease enzymes catalyze hydrolysis reactions to remove the RNA primers!
Similarly, eukaryotes and prokaryotes utilize different exonucleases! Eukaryotes primarily utilize RNase H while prokaryotes use DNA polymerase I as it actually has exonuclease activity.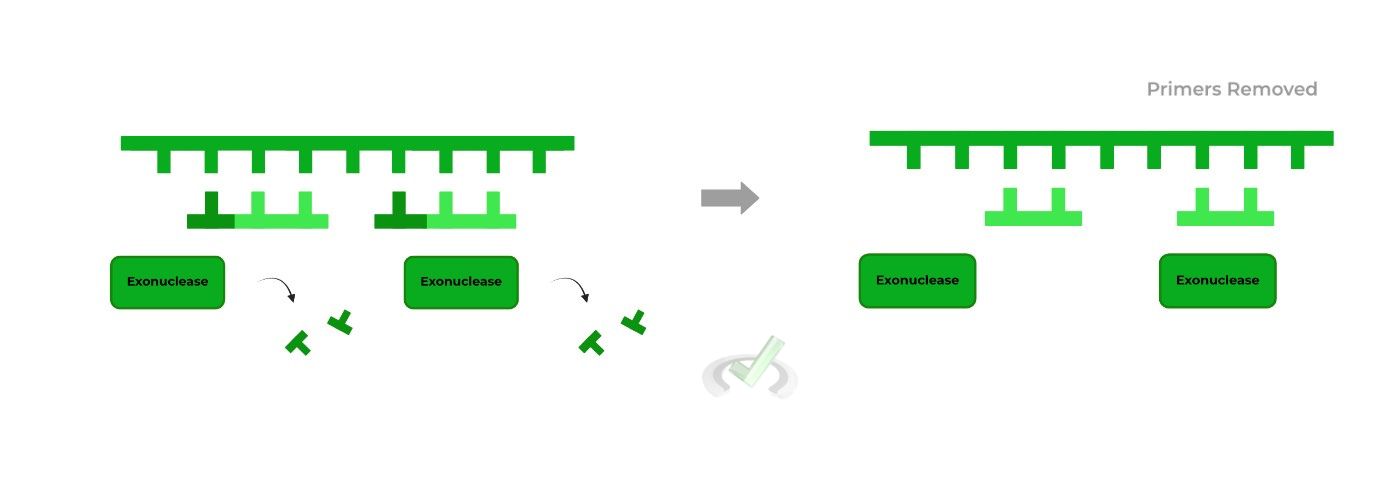
IV. DNA Ligase
Falling into the ligase class of enzymes (shocker!!), DNA ligase is highly important in the context of lagging strands and Okazaki fragments.
Recall that the lagging strand is composed of Okazaki fragments due to it being discontinuous. In order to synthesize a complete, intact DNA strand, these fragments must be “ligated” together, which is the function of DNA ligase.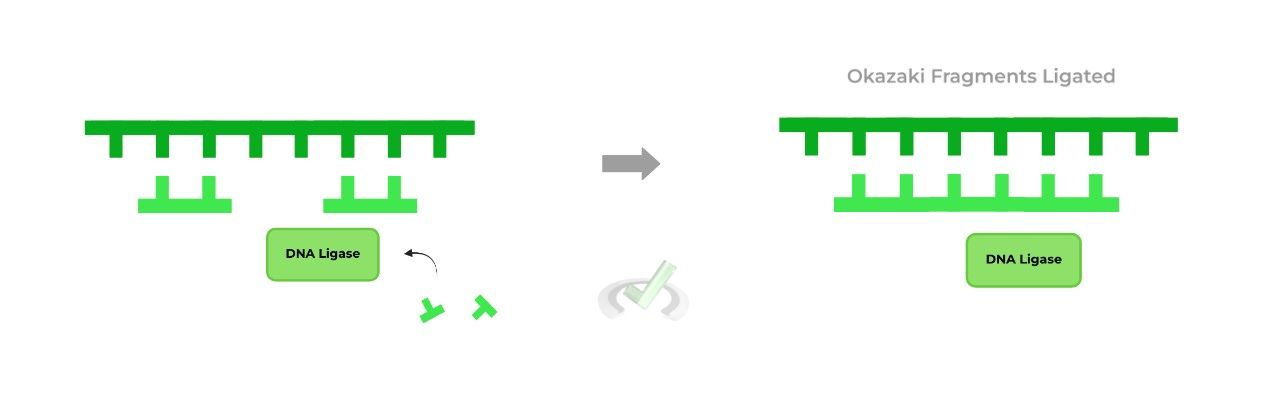
III. Bridge/Overlap
We have to give a 2nd apology in this article as we’ll have to give a quick call back to some basic organic chemistry concepts (we’re so sorry!). But, we’ll try to keep it as simple as possible!
I. Nucleophilic Substitution Reaction: Specific Coupling of Nucleic Acids
As stated above, DNA polymerase III catalyzes the addition of nucleotides to the growing DNA strand through a nucleophilic substitution reaction, the same reaction that occurs during peptide bond synthesis!
Take a look at the mechanism below and see how the reaction is applied in a biochemical context like this! Can you identify the nucleophile in this scenario?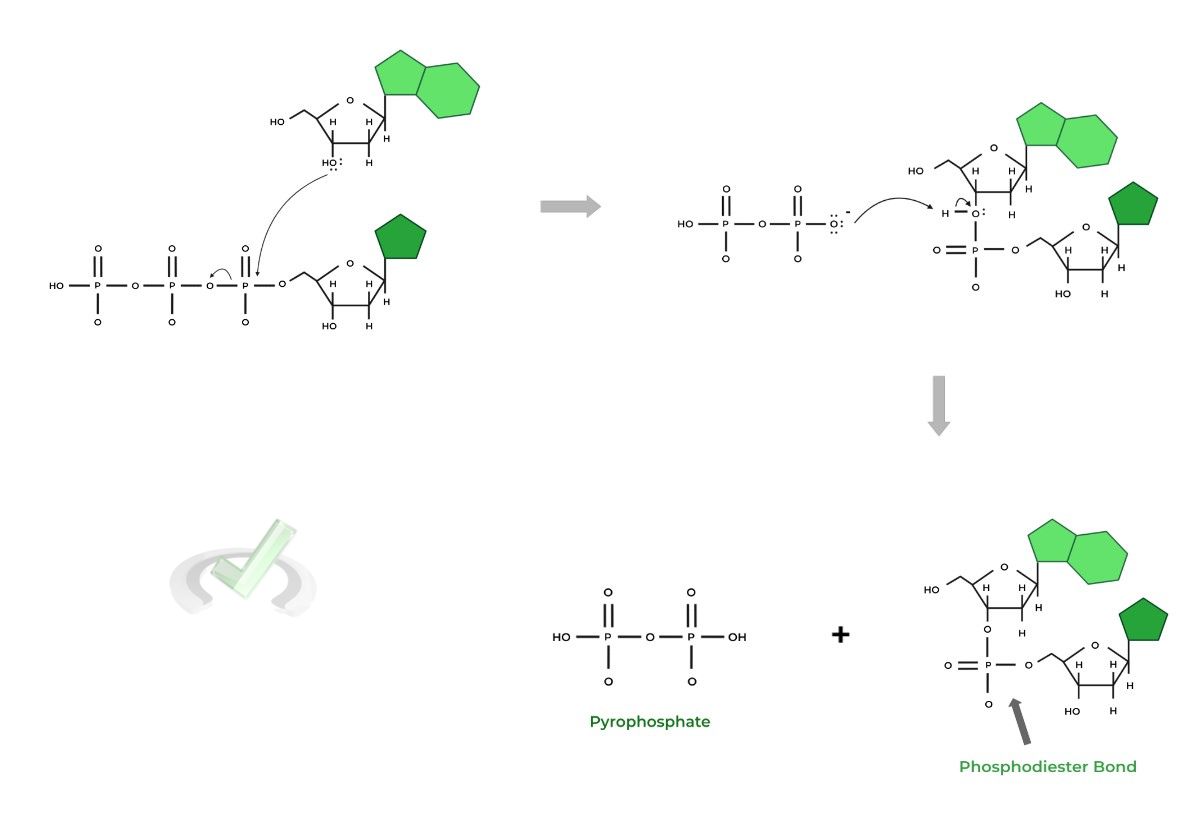
You’re absolutely right if you thought it said it was the C3 hydroxyl group! This is actually important for the coupling of incoming nucleotides as nucleotides will only be added to the chain when the C3 nucleophilic hydroxyl group is present!
Additionally, this will also be important when you review Sanger sequencing and the specific mechanism behind the experimental technique!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. General Mechanism of Replication Overview
This will be a little different as we’ll really just recommend and refer you to go back to the diagram earlier in the article as it already has a fair overview of the main enzymes in DNA replication!
B. Specific Enzymes Involved in Replication
Though all the above mentioned enzymes are important to DNA replication, we’ll go more in depth with: Primase, DNA Polymerase, Exonuclease, and DNA Ligase.
I. Primase
A specialized type of RNA polymerase that lays out the RNA primers. Interestingly, eukaryotic cells need these RNA primers as DNA polymerases have not evolved to initiate DNA strand synthesis without the need for a RNA primer.
II. DNA Polymerase
Probably the most important enzyme involved in DNA replication as it allows for the synthesis of the DNA daughter strand via a nucleophilic substitution reaction!
Recall that DNA polymerase can only read template strands in the 3’-5’ direction, which is why DNA strand synthesis only proceeds in the 5’-3’ direction.
Eukaryotes will utilize a combination consisting of DNA polymerase 𝛼, 𝛿, and 𝟄 while prokaryotes use DNA polymerase III.
III. Exonuclease
These enzymes remove the RNA primers in order to ensure that only DNA is present within the daughter strands.
Eukaryotes primarily utilize RNase H while prokaryotes use DNA polymerase I as it actually has exonuclease activity.IV. DNA Ligase
As indicated by its name, this enzyme ligates the Okazaki fragments on the lagging strand. Before DNA ligase can enact its function, recall that the RNA primers must first be removed by the exonuclease enzymes.
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
DNA polymerase III catalyzes the formation of what type of bond between nucleotides?
A. Phosphodiester Bond
B. Ionic Bond
C. Hydrogen Bond
D. Electrostatic Bond
Ans. A
DNA polymerase III catalyzes the lengthening of the DNA daughter strand utilizing a nucleophilic substitution reaction. This results in the phosphodiester bond which ends up forming the DNA phosphate backbone.
Sample Practice Question 2
Would the function of the ssDNA binding proteins come before or after the function of DNA helicase?
A. Before; because the DNA will have already been unwounded by then.
B. Before; because DNA helicase will bind to the single stranded DNA to prevent reassociation.
C. After, because DNA helicase will first unwind the DNA double helix.
D. After, because DNA gyrase works to unwind the DNA.
Ans. C
ssDNA binding proteins should work AFTER the function of DNA helicase because DNA helicase will first work to unwind the DNA double helix, exposing the single DNA strands.
Afterwards, the exposed single-stranded DNA will be bounded by the ssDNA binding proteins to prevent reassociation.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these