I. What are Key Terms in DNA Replication?
Imagine you’re a QB trying to run a pass play while playing football during a practice scrimmage with the hopes of getting the touchdown. How much harder do you think it’d be to run this play if you didn’t know what a cover 2 defense is or the proper running route for a hitch & go?
Even if you don’t play or are familiar with football, hopefully the analogy remains the same when discussing DNA replication! When we mention key terms, we’re talking about terms that can be used to generally describe DNA replication without getting into all the enzyme names and functions.
We actually believe that learning these key terms first in regards to DNA replication can actually become the basis and foundation for understanding the functions of the associated enzymes involved in DNA replication.
II. Fundamentals of DNA Replication
Before getting into the actual molecule itself, let’s first look at DNA replication at a more wide-scale view, specifically talking about the term, semiconservative.
A. Semiconservative Nature of Replication
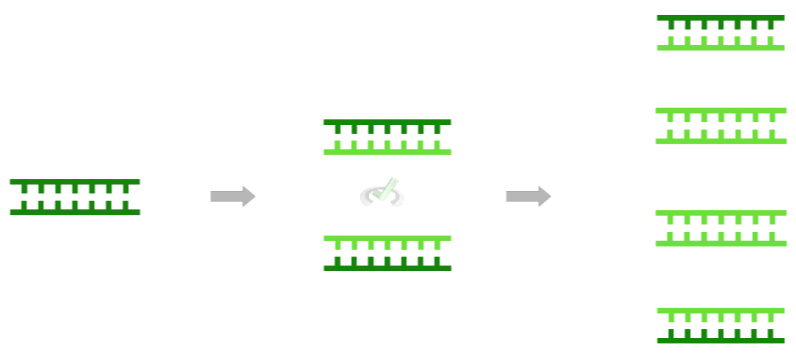
Because DNA is a complementary, double-stranded molecule,, the 2 strands can actually be used as templates, also termed the parent strands, in order to synthesize the new strands, termed the daughter strands, during replication.
Here, the term semiconservative refers to the fact that during each round of DNA replication, the parent (original) strands are still preserved.
However, as shown in the figure below, the parent strands slowly become outnumbered by the daughter strands, as their percentage contribution to all the DNA molecules exponentially decreases!
B. Origins of Replication and Replication Forks
These are exactly what they sound like: sequences, or regions, located on the DNA molecule which DNA replication proteins recognize in order to initiate replication!
We’ll cover all the enzymes involved in more detail in another article, but one enzyme we wanted to mention here is helicase, as the enzyme loads into these origins and unwinds the DNA to separate the strands!
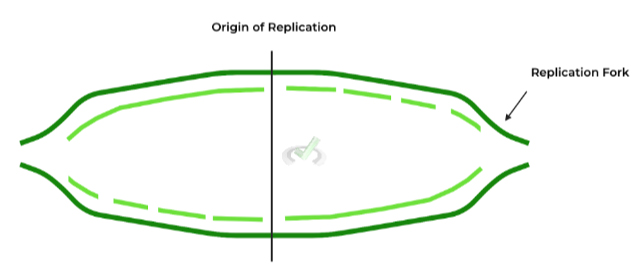
The additional term on the figure is the replication fork, which is used to describe the structure that’s formed from DNA unwinding. It’s easier to think of it as a shape, as it kinda looks like a 2 pronged fork!
I. Leading v.s. Lagging Strands
This is a topic that’s a little more complex but nevertheless a high yield one. As such, we’ll try to break down the topic as simple as possible! Additionally, this concept also helps you understand how certain DNA replication enzymes function!
Recall that DNA is arranged via an antiparallel orientation, where one end will be 3’ and the other will be 5’.
It’s important to understand that daughter strands are ONLY synthesized in the 5’ to 3’ direction. As such, the starting nucleotide on the template strand must be 3’ due to the antiparallel orientation.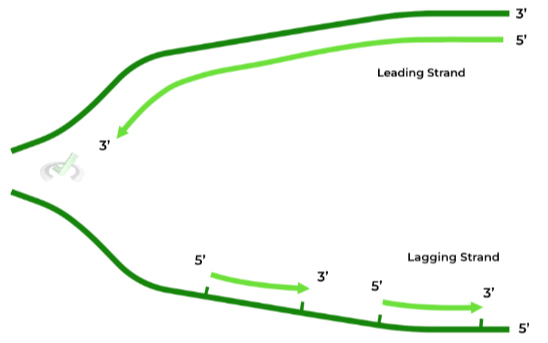
This becomes a problem as one of the template strands has a 5’ end. In this case, the daughter has to be made in little “chunks” or “fragments”, specifically called Okazaki fragments.
Furthermore, the daughter strands have specific names depending on whether it’s synthesized continuously or requires the use of fragments as shown above:
The leading strand is also termed the continuous strand, as it can be synthesized straight through without the need of any fragments, as its template strand has a starting 3’ end.
The lagging strand is also termed the discontinuous strand, as it CANNOT be synthesized straight, in which it does require fragments in order to synthesize.
Another way to think about their differences is that leading strand synthesis occurs “without interruptions” while lagging strand synthesis occurs “with interruptions”!C. Telomeres: Replicating the Ends of DNA Molecules
Another key concept to memorize is that after every round of replication, the amount of DNA on the daughter DNA molecules actually DECREASES, which poses a problem as at a certain point, the DNA coding genetic information will be lost!
To combat this potential loss of genetic information, our cells have evolved telomeres, which are repeating, noncoding sequences located on the ends of DNA, with the specific sequence being: 5’-TTAGGG-3’.
These will be the sequences that will be lost after a round of replication; however, because they are noncoding sequences, no genetic information is lost!
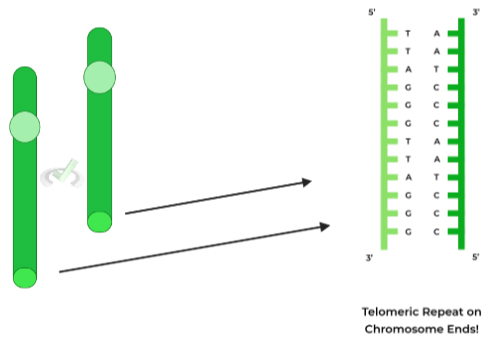
Think about the telomeres as if they’re “capping” the ends of DNA, similar to an aglet on a shoelace! (s/o to you if you thought about that one Phineas and Ferb song
III. Bridge/Overlap
DNA replication definitely has varieties among the different domains of life. One important difference occurs between prokaryotes and eukaryotes. Let’s take a closer look at this main difference!
I. Single v.s. Multiple Origins | Prokaryotes v.s. Eukaryotes
As stated above, one of the main differences between prokaryotic and eukaryotic DNA replication is that the former houses a SINGLE origin of replication while the latter houses MULTIPLE origin of replications.
Why does this difference occur? It actually comes down to the difference between how the DNA is organized between prokaryotes and eukaryotes.
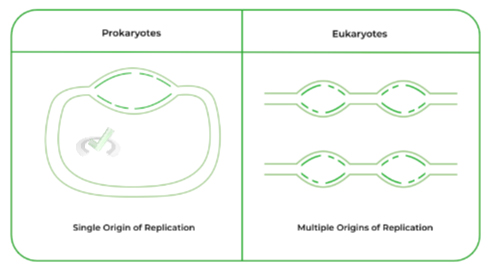
Recall that prokaryotes don’t have a “true nucleus” and instead have circular DNA. In contrast, eukaryotic DNA is organized via chromosomes which are condensed structures of linear DNA.
Because eukaryotic DNA is much larger and linear, having only 1 origin of replication would be time inefficient for DNA replication. As such, multiple origins of replication are used in order to speed DNA replication for eukaryotic DNA!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Semiconservative Nature of Replication
One general term to describe DNA replication is semiconservative because the parent (template) strand from the original DNA molecule is always preserved in the new DNA molecules!
However, with each round of DNA replication, the parent strands slowly become outnumbered by the daughter (new) strands!B. Origins of Replication and Replication Forks
Origins of replication are DNA sequences/regions that can be identified by DNA replication proteins in order to initiate replication.
One important DNA replication enzyme that has a major role in the origins of replication is helicase, which works to unwind the DNA strands at the origin.
The replication fork is used as a term to describe the structure/shape that’s formed due to the unwinding of DNA during replication. As the name implies, the shape that’s made kinda looks like a two-pronged fork when the DNA is unwinding!I. Leading v.s. Lagging Stands
An important concept to understand when learning about leading & lagging strands is that daughter strands are ONLY synthesized in the 5’ to 3’ direction. As such, there are 2 types of daughter stands:
Because the leading (continuous) strand has a template strand starting with a 3’ end, it can replicate straight through “without interruptions” and does not require the use of fragments.
Because the lagging (discontinuos) strand has a template strand starting with a 5’ end, it CANNOT replicate “straight through” and does require the use of Okazaki fragments.C. Telomeres: Replicating the Ends of DNA Molecules
Because the amount of DNA is lost after every round of replication, eukaryotic cells use telomeres, which are repeating, noncoding sequences that act as a “cap” to prevent the loss of genetic information after every round of replication.
Specifically, the repeated nucleotide sequence is 5’-TTAGGG-3’ and actually came about as a result of evolution!V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
Given the figure shown below, depicting two strands of DNA separated, where would you expect the leading and lagging to be located, respectively?
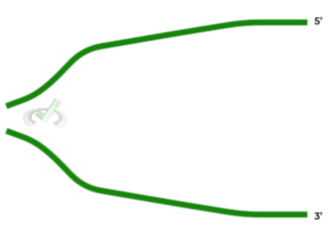
A. Top; Bottom
B. Bottom; Top
C. Top; Top
D. Bottom; Bottom
Ans. B
Luckily with this question, you can already take away C and D as answers. Recall that the leading strand is continuous and will have a template strand with a starting 3’ end. Conversely, the lagging strand is discontinuous and will have a template strand with a starting 5’ end.
As such, this is why the lagging strand requires the use of Okazaki fragments in order to complete daughter strand synthesis.
Sample Practice Question 2
Given a starting DNA molecule, what is the percentage of the PARENT DNA strands that make up the DNA molecules after 2 rounds of replication?
A. 0%
B. 25%
C. 50%
D. 100%
Ans. B
Recall that after every round of DNA replication, the parent (original) DNA strands slowly start to become outnumbered by the daughter (new) DNA strands. Let’s do the math real quick!
After 1 round, there will be 2 daughter strands and 2 parent strands ⇒ The percentage of the parent strands is 50% (2/4 → 1/2).
After another round, there will be 6 daughter strands and 2 of the original parent strands ⇒ The percentage of the parent strands is 25% (2/8 → 1/4)





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these 










 reviews on TrustPilot
reviews on TrustPilot