I. What is DNA Sequencing?
It’s amazing to think that with all the modern innovations and breakthroughs in biotechnology that we’ve been able to sequence the entire human genome and the genomes of many other organisms! Just goes to show that molecular biology has come a long way!
Aside from the human genome, DNA sequencing has been such a beneficial and groundbreak laboratory technique when it comes to research of all kinds from immunology, cancer biology, pathophysiology and so much more!
Fortunately, DNA sequencing builds off of some of the fundamental principles that drive DNA replication and even gel electrophoresis, with some caveats twisted in! We’ll make sure to connect all of these together as concisely as possible below, let’s get started!
II. DNA Sequencing
DNA sequencing allows us to determine the nucleotide sequence of a gene. When it comes to the MCAT, a technique called Sanger sequencing, particularly its modern day variation. Let’s take a look at them below!
A. Overview of DNA Sequencing
This technique was developed by Dr. Frederick Sanger, most notable for its use of dideoxyribonucleotides and polymerase chain reactions in the process.
I. Dideoxyribonucleotides
An important component of Sanger sequencing is the incorporation of dideoxyribonucleotides in the reaction, also termed chain terminating nucleotides. The figure below compares the structures between dNTPs and ddNTPs. (Note: nucleoside version is shown for simplicity)
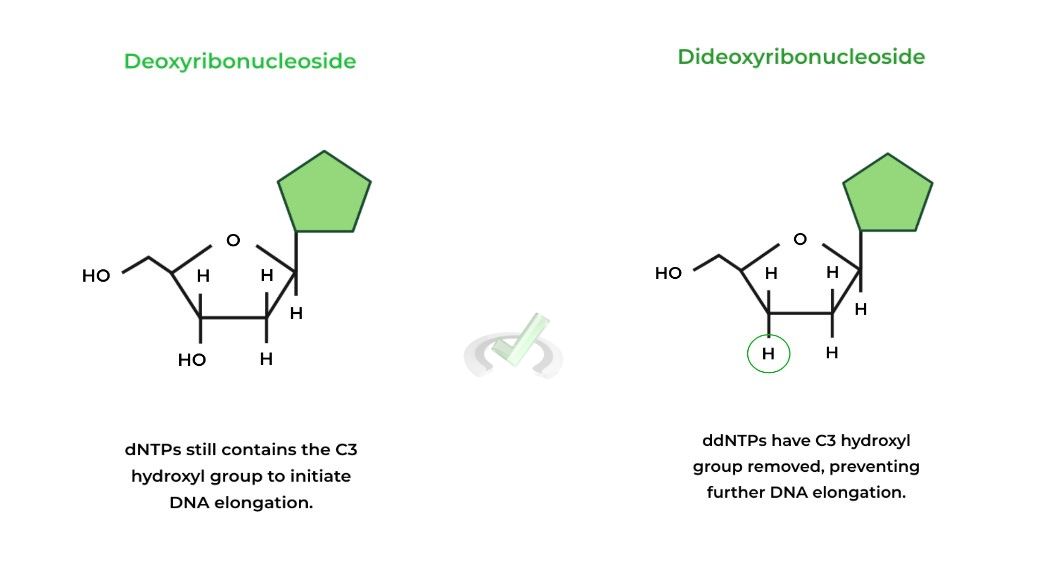
Note that both can have A, T, C, and G as their nitrogenous base and also have the triphosphate group, but differ where ddNTPs have the C3 hydroxyl group removed, hence the term “-dideoxy”.
This is important because if a ddNTP is added to a growing DNA strand, further DNA elongation will stop as there is no C3 hydroxyl group to initiate the nucleophilic substitution which elongates the DNA strand!II. Polymerase Chain Reaction
Though we’ve covered PCR in more depth in another article, it is also utilized here to help sequence the DNA molecule!
In order for Sanger sequencing to be successful, we need to generate lots of DNA strands of different lengths, which is where PCR comes into play!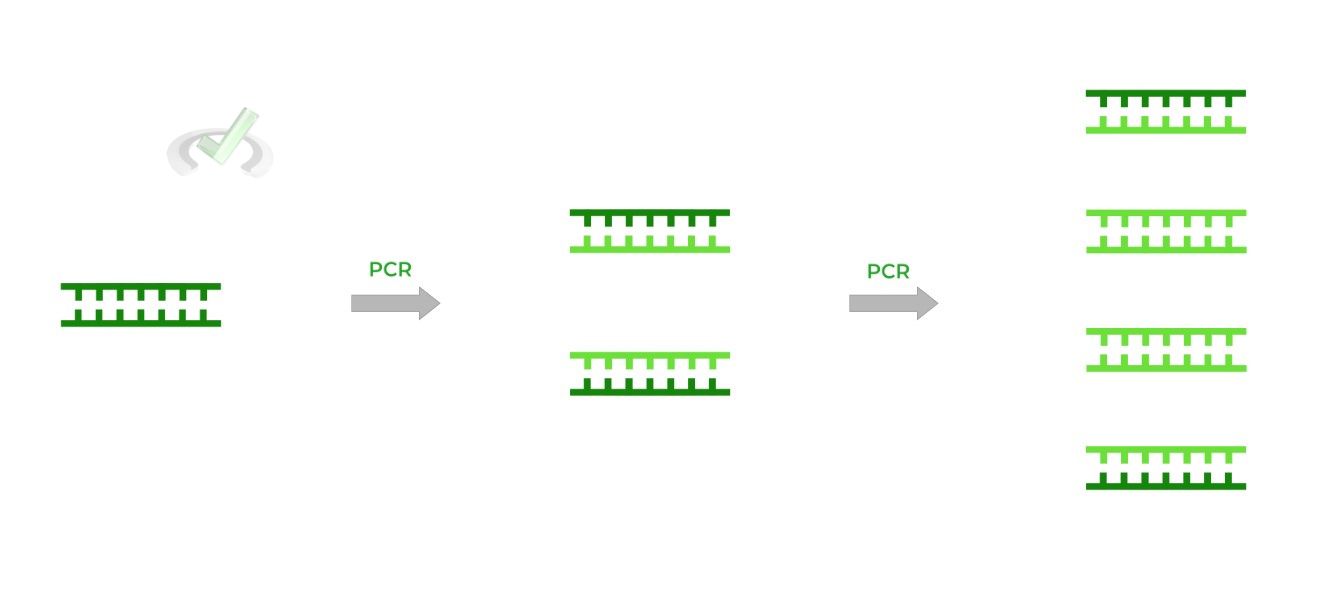
Because PCR allows us to generate DNA strands in a fast and efficient way, it’s combined with the ddNTPs terminating nucleotides to sequence the gene as we’ll explain below!
B. Sanger Sequencing: Process
In order to simplify the process as much as possible, we’ve identified 3 main steps that are involved in order to sequence the gene: 1) Template Stand Isolation, 2) PCR & Chain Termination, and 3) Gel Electrophoresis. Let’s cover them all in depth!
I. Template Strand Isolation
This first step is relatively simple, as we’re only trying to isolate the template strand that needs to be sequenced, which requires denaturation of the double DNA strand!
This is usually done via heating, similar to PCR, which breaks the hydrogen bonds and separates the 2 strands.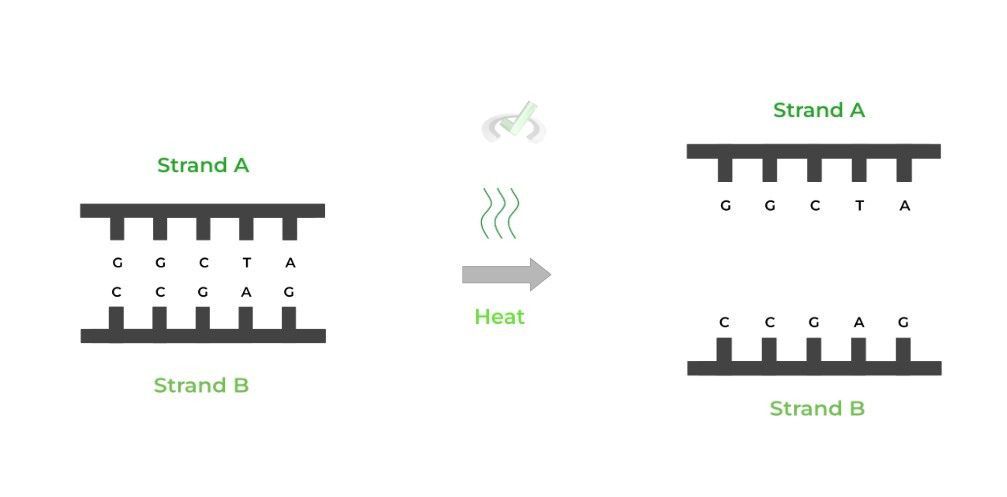
Note that because we don’t know which gene is coding and noncoding, 2 separate Sanger sequencing reactions will be performed for each strand.
II. PCR & Chain Termination
For this example, let’s say we’re trying to sequence strand B. A primer is now attached in order to initiate elongation. Recall that DNA polymerase can only add nucleotides to an existing strand of nucleotides.
The template strand and primer are then included in a test tube which contains deoxyribonucleotides (dNTPs) and dideoxyribonucleotides (ddNTPs), where the dNTPs are in a higher concentration.
Additionally, the ddNTPs have a fluorescent label attached where each base has a different color; for example, thymine might have a dark green while cytosine has a teal. Now, let’s run 1 PCR to see what happens!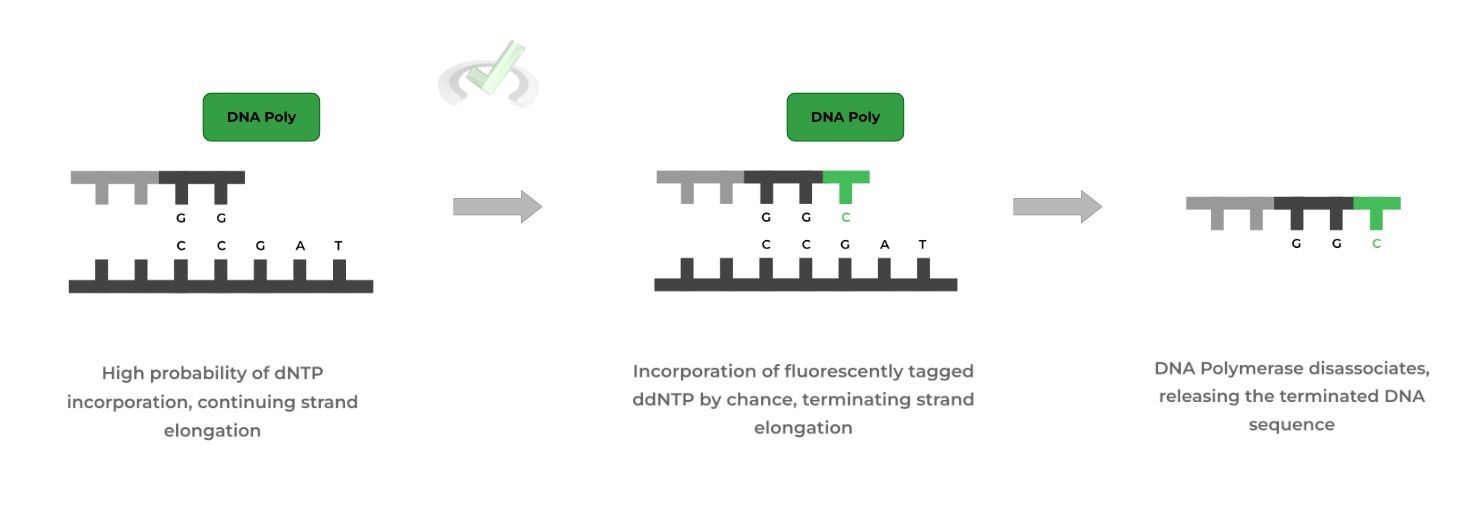
When the PCR is run, mostly dNTPs will be incorporated and will continue to allow for chain elongation because the C3 hydroxyl group is still present!
However, every now and then, one of the uniquely colored ddNTPs will be incorporated. Because they don’t have the C3 hydroxyl group, the DNA strand will stop growing resulting in chain termination!
After many cycles, you'll accumulate many DNA strands of different lengths and more importantly, different chain terminating ddNTPs, as shown below! (Note: primer not shown).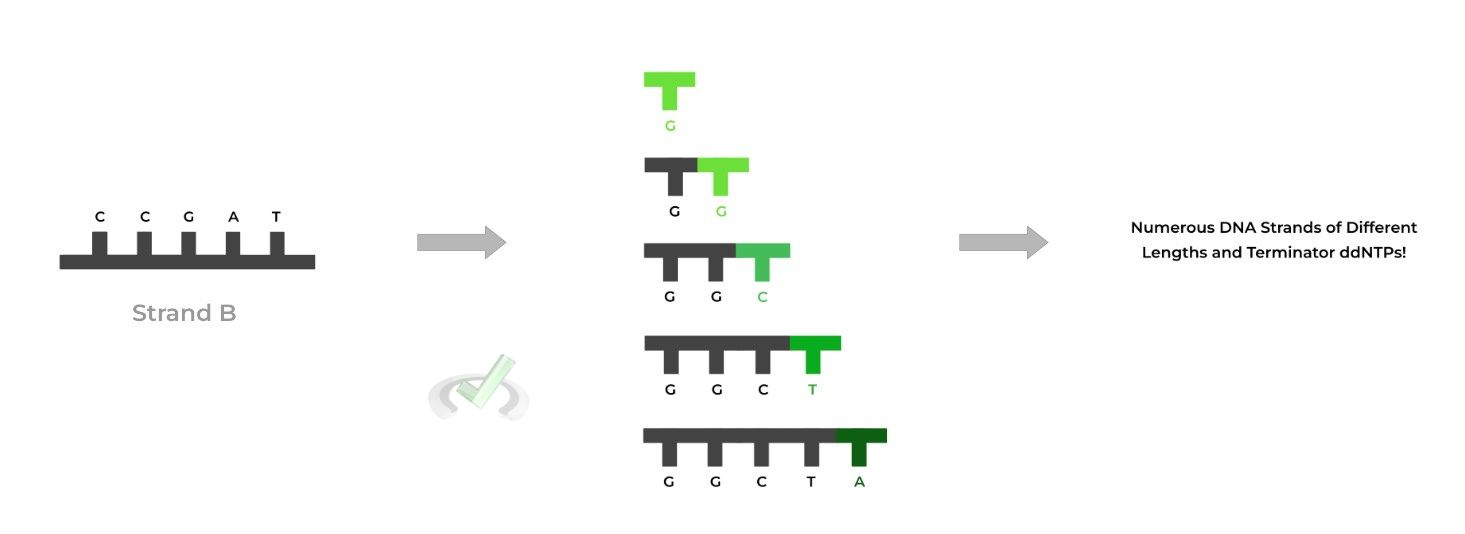
III. Gel Electrophoresis
In a previous article, we looked at utilizing gel electrophoresis in the context of proteins. Now, we’re doing it with DNA stands to help identify the sequence!
Recall that gel electrophoresis can separate molecules via size where larger molecules will travel a smaller distance and smaller ones will travel farther.
We now have a lot of DNA strands with different lengths and terminator ddNTPs; now, these DNA strands can be run on a gel and be separated via size!
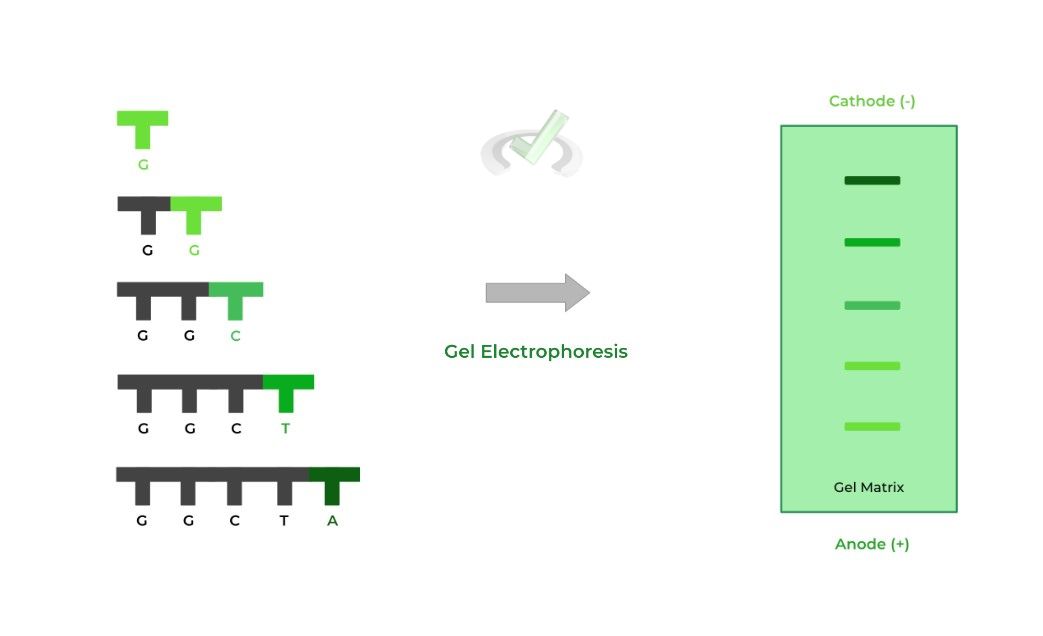
There are 2 things to consider now when determining the sequence: 1) the distance traveled by the molecule (i.e. band) and 2) the color of the fluorescent tag.
Because the longest DNA strand will travel the least distance, this indicates that the terminator ddNTP is the LAST nucleotide of the gene, and thus will be on the 3’ end.
Likewise, the shortest DNA strand will travel the farthest, indicating that the terminator ddNTP is the FIRST nucleotide of the gene, and thus will be on the 5’ end.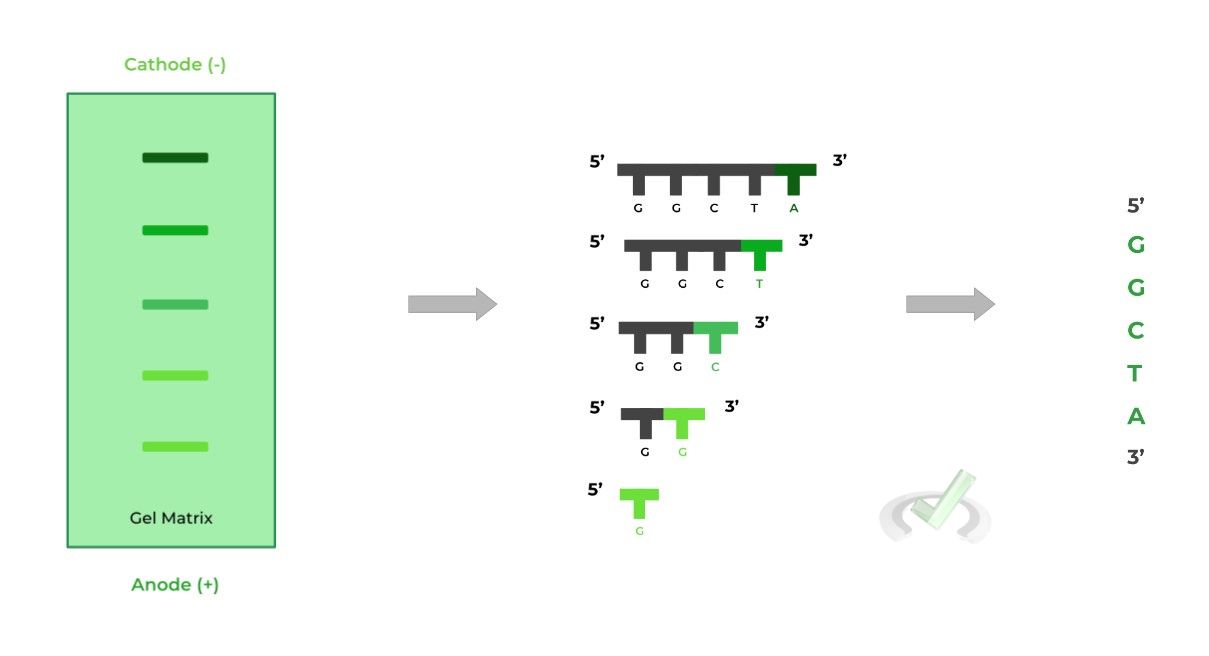
This is also where the uniquely colored fluorescent tags are helpful! Because each ddNTP base has a unique fluorescent tag color, we’ll be able to know which base is the one that terminated the chain!
One way to think of it is that the fluorescent tag allows us to determine the nucleotide while gel electrophoresis allows us to determine its position! Now you’ve determined the nucleotide sequence!
III. Bridge/Overlap
Recall that the addition of dNTPs (and ddNTPs) can be thought of in an organic chemistry reaction, specifically a nucleophilic substitution! Let’s quickly review the reaction below!
I. Nucleophilic Substitution in DNA Elongation
Recall that in a nucleophilic substitution, a nucleophile is generally an electron dense species that attacks an electrophile in order to replace a functional group attached to the electrophile.
In the context of nucleotide addition, the C3 hydroxyl group functions as the nucleophile attack the electrophilic phosphate of the 𝛼- phosphate group!
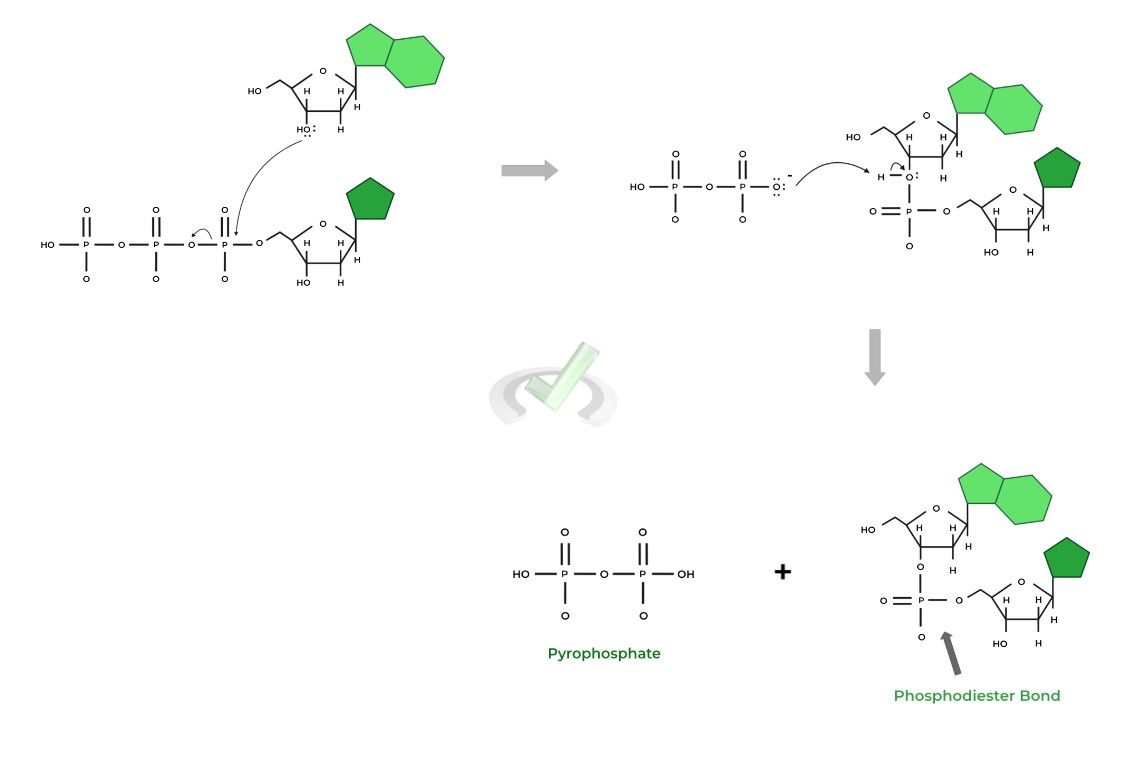
Hopefully now it’s a little clearer as to why the dideoxyribonucleotides terminate the growing DNA chain! It’s because they lack the C3 hydroxyl group which acts as the nucleophile to catalyze the formation of another phosphodiester bond!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Overview of DNA Sequencing
Sanger sequencing, developed by Dr. Frederick Sanger, is most notable for its use of dideoxyribonucleotides and polymerase chain reactions in the process.
I. Dideoxyribonucleotides
These nucleotides are very similar to the more familiar deoxyribonucleotides; however, they differ where the dideoxyribonucleotides (ddNTPs) lack the C3 hydroxyl group, hence the term “-dideoxy”.
Because of this, when attached to a growing DNA strand, elongation will terminate as there is no C3 hydroxyl group to catalyze the addition of another nucleotide. This is why they’re also called terminator nucleotides.
II. Polymerase Chain Reaction
In order for Sanger sequencing to be successful, we need to generate multiple copies of DNA, each with different lengths and terminator ddNTPs.
We can use multiple polymerase chain reactions to generate these multiple DNA strands of different length and terminator ddNTPs.
B. Sanger Sequencing Process
In order to simply the process as much as possible, we’ve identified 3 main steps involved in Sanger sequencing as listed below:
I. Template Strand Isolation
In order to isolate the strand of DNA to be sequenced, heat is first applied to the dsDNA molecule in order to break the hydrogen bonds and separate the strands.
II. PCR and Chain Termination
The template strand is then included in a test tube also consisting of a primer, DNA polymerase, dNTPS, and ddNTPs.
It’s important to note that there is a higher concentration of dNTPs compared to ddNTPs in the mixture. In addition, the ddNTPs are also fluorescently tagged, each base having a unique color.
After multiple cycles of PCR, we’ll have DNA strands of varying lengths as well as varying terminator ddNTPs. They’re then run under gel electrophoresis!
III. Gel Electrophoresis
Remember that gel electrophoresis can separate molecules by size, where large molecules travel short distances and small molecules travel further.
In this case, the largest strand will run the least distance, meaning that the terminator nucleotide (determined by the fluorescent color) will be the LAST of the sequence and will be on the 3’ end.
Likewise, the smallest strand will run the farthest distance, meaning that its terminator nucleotide will be the FIRST of the sequence and will be on the 5’ end.
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
Given the following gel of the DNA strands after multiple PCR cycles, what is the first nucleotide of the gene sequence?
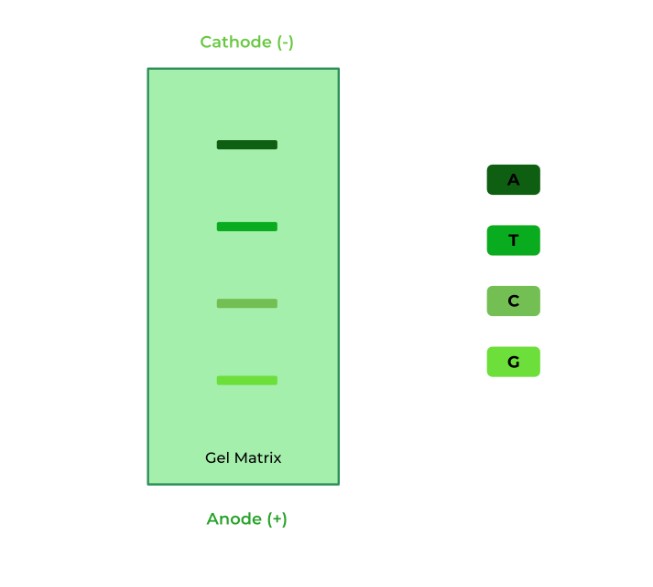
A. A
B. T
C. C
D. G
Ans. D
Recall that the smallest DNA strand will travel the farthest, and thus will be the FIRST nucleotide of the sequence located on the 5’ end! Try and reconstruct the DNA strands of varying lengths and see if that helps with the rationale!
Sample Practice Question 2
Given the following gel, which of the following is the correct sequence?
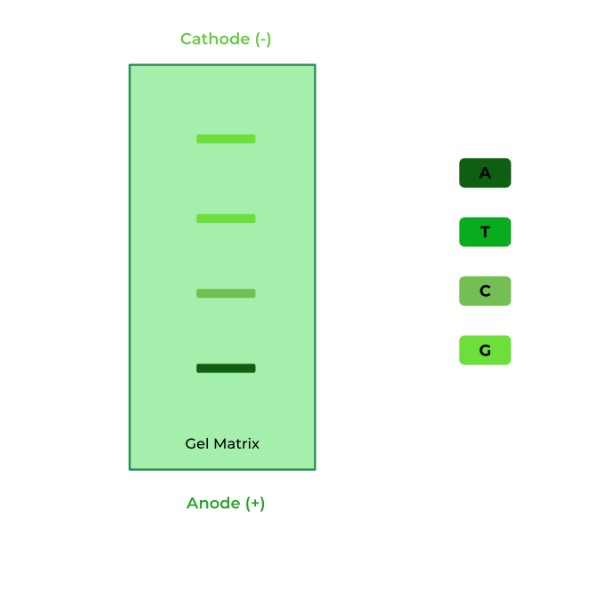
A. 5’-ACGG-3’
B. 5’-GGCA-3’
C. 5’-TCTG-3’
D. 5-GTCT-3’
Ans. A
Remember always that the smallest DNA strand will travel the furthest, meaning that its terminator nucleotide is the FIRST nucleotide of the sequence located on the 5’ end.
Conversely, the largest DNA strand will travel the least distance, meaning its terminator nucleotide is the LAST nucleotide of the sequence, located on the 3’ end.
As shown, A is the farthest that travels, meaning that it will be the FIRST nucleotide, on the 5’ end. G travels the least distance, meaning that it will be the LAST nucleotide, on the 3’ end.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these 










 reviews on TrustPilot
reviews on TrustPilot