I. What is Prokaryotic Gene Expression?
Nowadays, with our hectic, ending schedule especially for you guys as premeds, it’s definitely important to maintain a sense of balance between school, family, friends, extracurricular activities. There’s a right time for everything and finding that balance is important for all aspects of life!
Just like us, prokaryotes also need that balance in their microcellular life just in a less social sense. These organisms need to regulate what type of proteins to produce and when so as to maximize their efficiency within their lifespan.
In a previous article, we’ve already discussed and covered the basics of the central dogma, particularly RNA transcription. Now, we’re just applying what we learned in those lessons in a more applicable sense in terms of how that transcription is regulated!
II. Transcriptional Regulation in Prokaryotes
Before getting into the main topic on prokaryotic gene expression, let’s backtrack real quick to understanding a key difference between eukaryotic and prokaryotic transcription: monocistronic v.s. polycistronic genes!
A. Monocistronic v.s. Polycistronic Genes
Recall that one major difference between prokaryotic and eukaryotic transcription is that eukaryotes are monocistronic and prokaryotes have the ability to be polycistronic.
Monocistronic refers to the mRNA transcript ONLY coding for ONE protein while polycistronic means that the mRNA can code for MULTIPLE proteins!
B. Operon Concept and Structure: The Jacob Monod Model
The operon model of prokaryotic gene expression refers to the cluster of gene sequences and regions which work together to produce a mRNA transcript, probably better explained via the visual below.
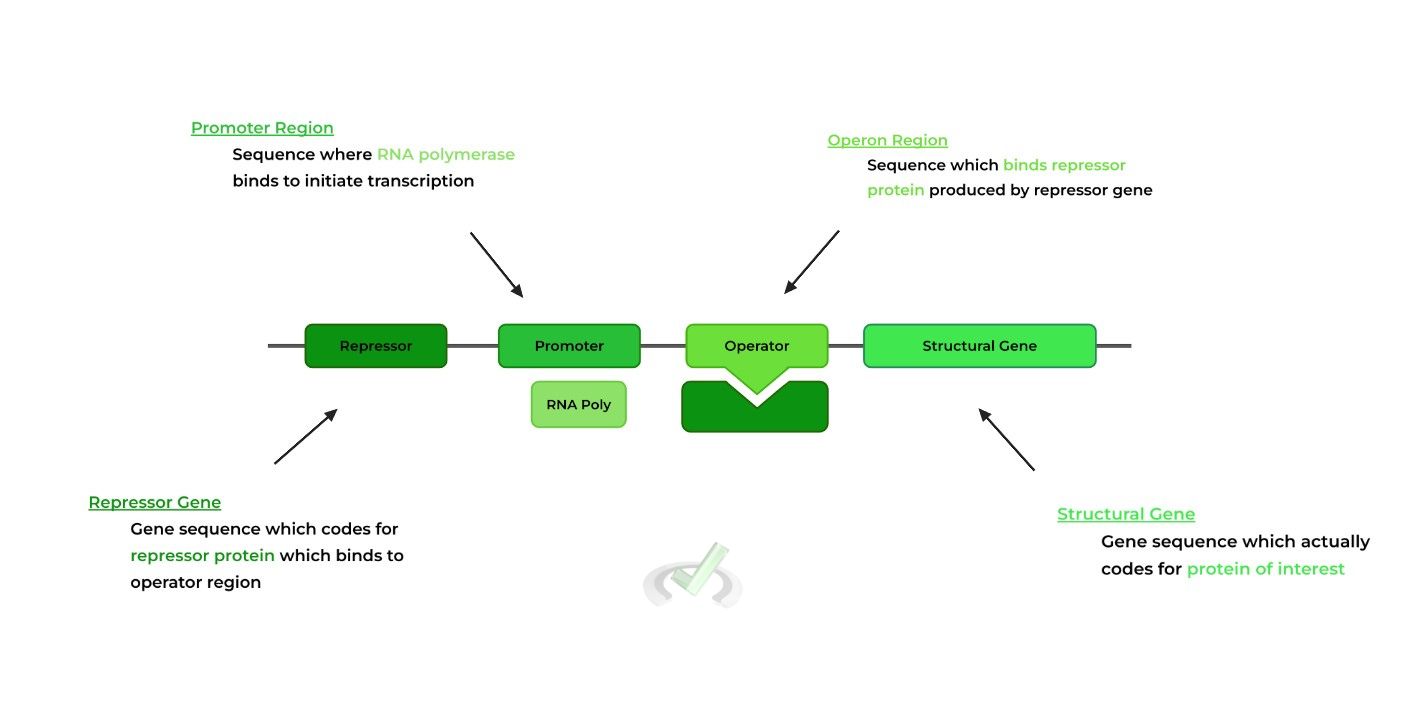
There are 2 genes within the model: the repressor gene, which codes for a repressor protein, and the structural gene, which codes for the actual protein of interest.
Between these 2 genes are the promoter region, which binds the RNA polymerase, and operator region, a sequence which binds the repressor protein.I. Importance of Sequence Order in Operon
The main takeaways with the operon model are the order of the sequences in the operon and the important interaction between the repressor protein and operator region.
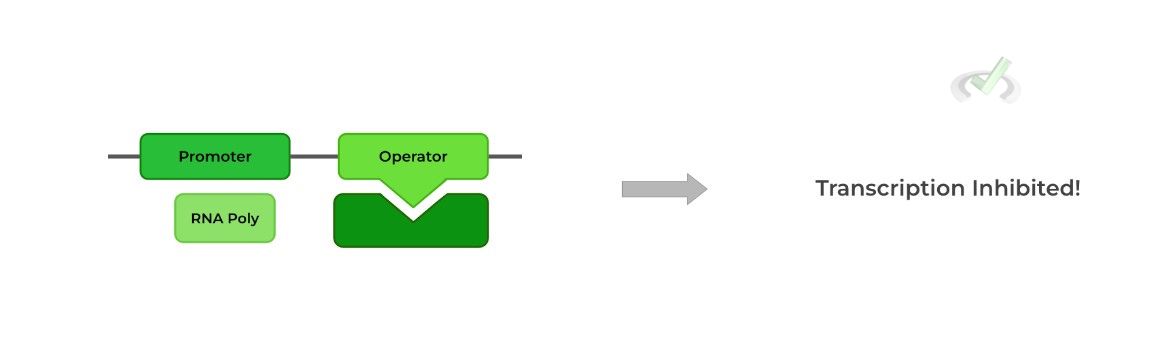
Because the operator region comes AFTER the promoter region, RNA polymerase CANNOT transcribe the structural gene because the operator bound repressor protein blocks the RNA polymerase from transcription.
C. Transcriptional Regulation: Types of Operon Models
There are 2 main categories for operon models: 1) an inducible model and 2) a repressible model, which differ mainly on whether it is constitutively active or inactive. Let’s take a closer look at them!
I. Inducible Operon System: Positive Control in Bacteria
In an inducible operon system, transcription of the structural gene is CONSTITUTIVELY INACTIVE because the repressor protein is constantly bound to the operator region.
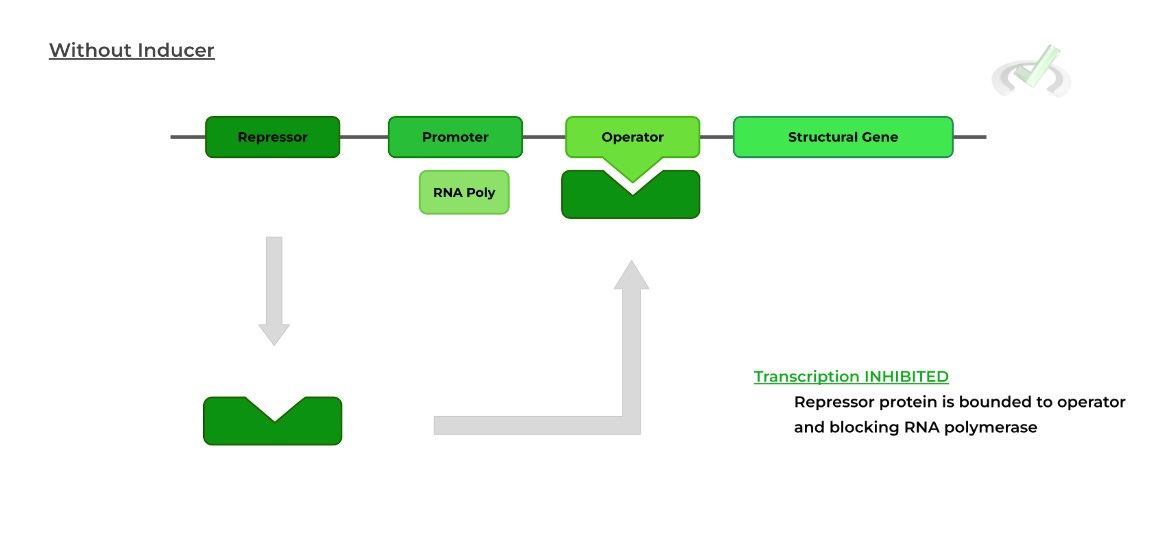
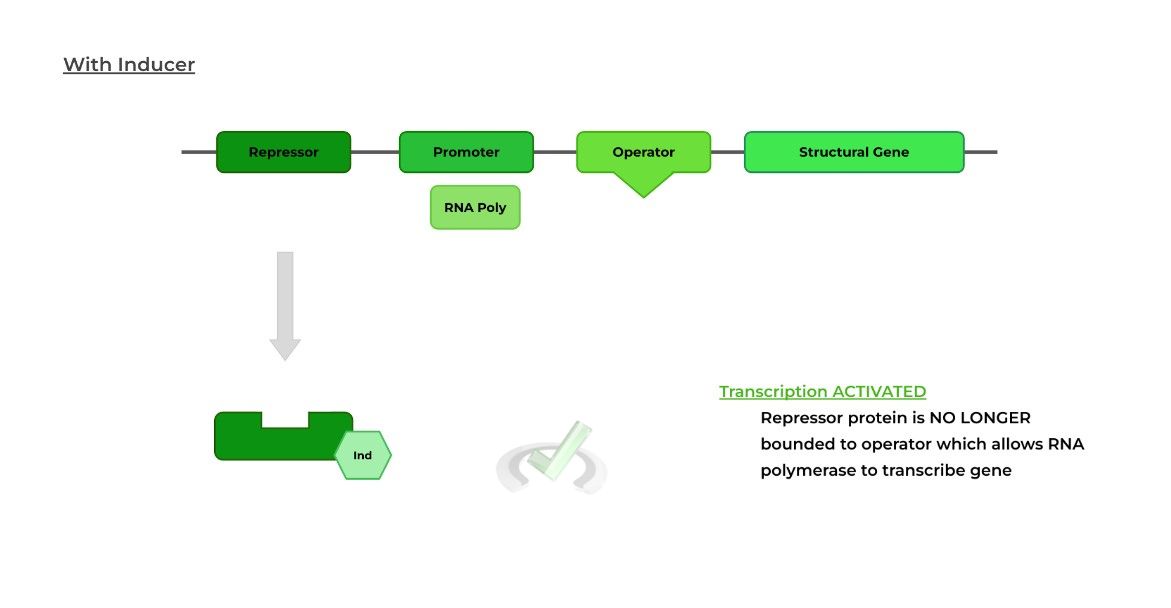
An outside agent or molecule binds to the repressor protein causing a change in expression. In this case, the external agent is called an inducer because it INDUCES gene transcription!
When the inducer binds to the repressor protein, a conformational change (triangle to square) occurs making the repressor unable to bind the operator.
Because the repressor protein is no longer bound to the operon, the RNA polymerase is no longer blocked and can proceed with transcription.II. Repressible Operon System: Gene Repression in Bacteria
In contrast, a repressible operon system is CONSTITUTIVELY ACTIVE transcription because the repressor protein is in a constant “inactive” state, where it can’t bind to the operator.
Here, the external agent is referred to as a corepressor because when it binds to the repressor protein, a conformational change occurs “activating” the repressor protein.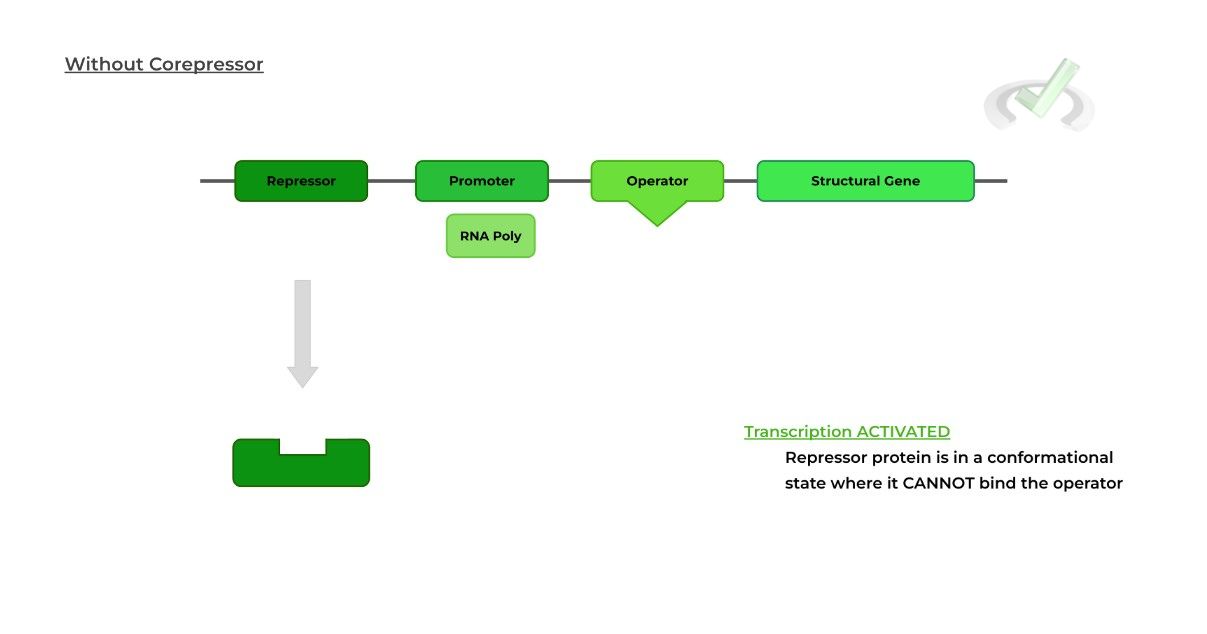
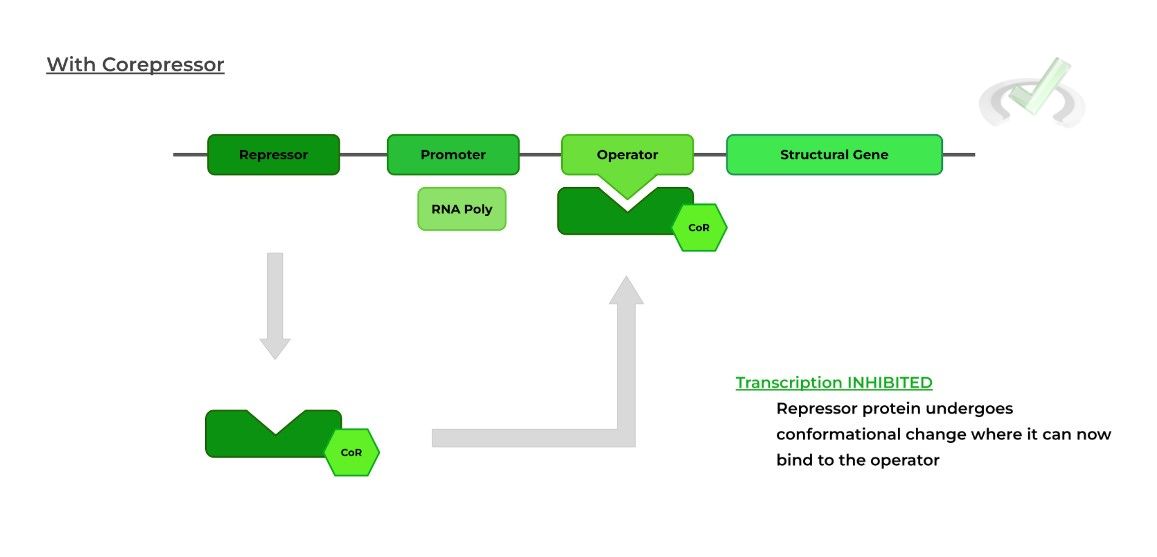
Likewise, in this scenario, the repressor protein switches from an “inactive'' state (square shape) to an “active” state (triangle shape).
Now, the repressor protein can bind the operator region and block the RNA polymerase from transcription.
III. Bridge/Overlap
This section, again, will be a bit more different and unique from other overlap sections as it’s more about developing a strategy and logic again!
I. The Lac Operon
As stated above, the best way to understand the operon models and their various systems is through real life examples implemented with the lifestyle of prokaryotes.
One example of an inducible system is the lac operon: this is actually also an example of a polycistronic gene, as it codes for multiple enzymes involved in lactate metabolism. In the case for the lac operon, the inducer is allolactose, an isomer of lactose.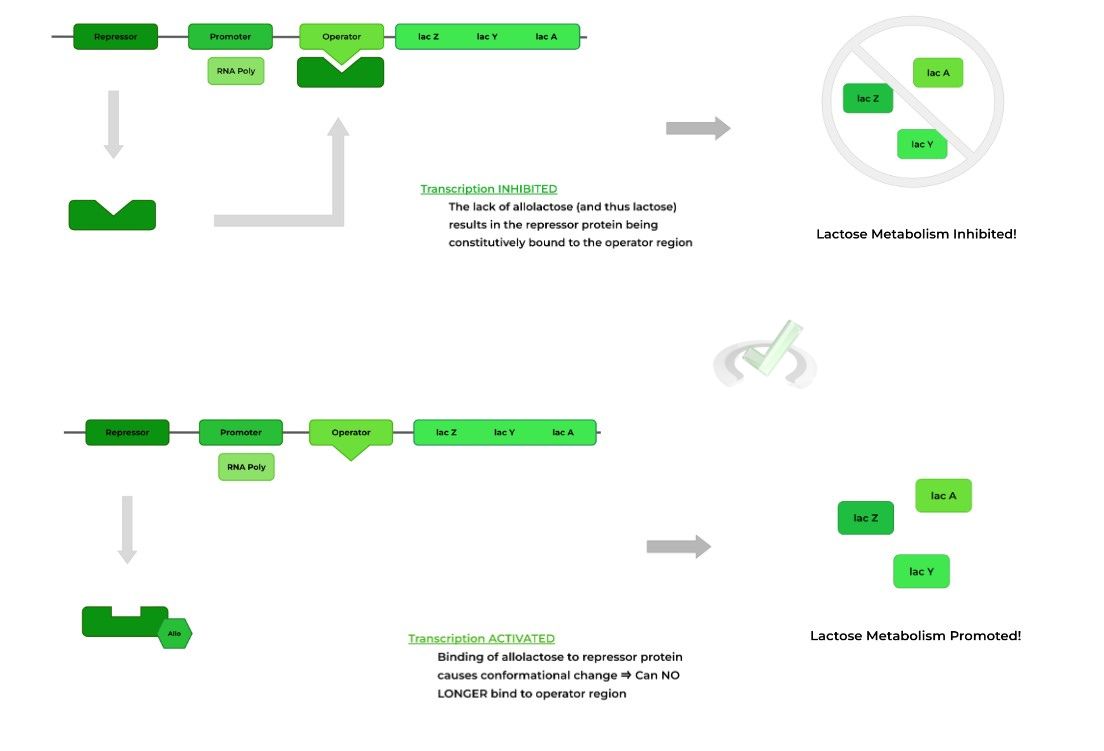
It’s beneficial and efficient for the prokaryote to only produce these lactate metabolizing enzymes ONLY in the presence of lactose!
If there were no lactose present, it would be a waste of energy and cellular machinery to transcribe and produce these enzymes.
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Monocistronic v.s. Polycistronic Genes
Eukaryotic genes are termed monocistronic because each mRNA transcript only codes for ONE protein. Prokaryotic genes are termed polycistronic because each mRNA transcript can code for MULTIPLE proteins!
B. Operon Concept and Structure: The Jacob Monod Model
One of the most widely seen ways prokaryotic gene expression is regulated is via the operon model! An operon contains 4 main genes/sequences:
The repressor gene codes for the repressor protein which binds to the operator region and the promoter is the sequence where RNA polymerase will bind to.
The operator region is the sequence where the repressor protein will bind to and the structural gene codes for the protein of interest.I. Importance of Sequence Order in Operon
The most important concepts to keep in mind about the operon are the order in which the sequences are placed and the interaction between the repressor protein and the operator region.
Because the operator region comes AFTER the promoter, the RNA polymerase cannot transcribe the gene when the repressor protein is bound to the operator region because it blocks the RNA polymerase. Hence, gene expression is halted.C. Transcriptional Regulation: Types of Operon Models
There are 2 main categories of operon models: 1) an inducible model and 2) a repressible model, which differ mainly on whether it is constitutively active or inactive.
I. Inducible Operon System: Positive Control in Bacteria
These systems are CONSTITUTIVELY INACTIVE because the repressor protein is constantly bound to the operator region.
Upon the binding of an inducing, exogenous agent, the repressor protein changes its conformation to where it CAN NO longer bind the operator region. As such, the RNA polymerase is no longer blocked and CAN proceed with transcription.II. Repressible Operon System: Gene Repression in Bacteria
These systems are CONSTITUTIVELY ACTIVE because the repressor protein is in a constant conformation where it cannot bind to the operator region.
Upon binding of a corepressing, exogenous agent, the repressor protein changes its conformation to where it CAN bind the operator region. As such, the RNA polymerase is now blocked and CANNOT proceed with transcription.V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
Suppose a mutation occurred to where the positions of the promoter and operator regions of the operon switched places. In this case, the gene would be constitutively active. Which of the following would aid in “turning off” the gene in this scenario?
A. Inducer
B. Corepressor
C. Inducer or Corepressor
D. None of the above.
Ans. D
This question comes down to understanding the importance of the ordering in the operon. In a regular operon, the operator region comes AFTER the promoter region; as such, when the repressor protein is bound to the operator, transcription is inhibited because the RNA polymerase is blocked.
In this case, the promoter comes AFTER the operator. No matter what exogenous agent is added, the gene will still be transcribed because the repressor protein is no longer blocking the RNA polymerase. As such, no exogenous agent can “turn off” the gene.
Sample Practice Question 2
The trp operon is a prokaryotic operon which codes for the enzymes that can synthesize tryptophan. It is shown that when there is an excess amount of tryptophan, transcription of the trp operon system is inhibited. Which of the following best describes tryptophan and the type of operon system it interacts with?
A. Inducible, Inducer
B. Corepressor, Repressible
C. Inducible, Corepressor
D. Repressible, Inducer
Ans. B
Of the following combinations, only A and B are valid pairings. As stated in the question stem, when there is an excess of tryptophan, the trp operon is inhibited. Likewise, we can consider tryptophan a compressor because it COREPRESSES the transcription of the gene by binding and “activating” the repressor protein.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these