I. What are the Processes of Translation?
Although the last part of the central dogma, translation can also be quite complex and intricate as DNA replication and RNA transcription, don't worry! We’ll make sure y’all don’t get “lost in translation” going through the article.
Corny jokes and puns aside, translation becomes a bit more easier to understand with the aid of visuals and diagrams as we’ll show below, just like replication and transcription! It also helps taking it a step at a time, which we’ll do our best to do in the article!
Just like before with DNA replication and RNA transcription, you’ve covered the basics, key terms, and fundamentals of translation as outlined in our previous article: now it’s all just a matter of applying them to the actual process!
II. Initiation, Elongation, and Termination of Translation
Before getting into the actual processes and steps of translation, let’s get a quick refresher of the ribosome structure.
A. Review of Ribosome Structure
Recall that the ribosomes are 3 main sites: the A (aminoacyl) site, P (peptide/peptidyl) site, and E (exit) site. Listed below are the main characteristics of each site:
The A (aminoacyl) site serves as the entry point for incoming tRNAs charged with amino acids. The P (peptide/petidyl) site is where elongation of the polypeptide chain occurs. The E (exit) site is where empty tRNAs can exit to be recharged with another amino acid.B. Process of Translation
The entire process of translation can actually be subdivided into 3 main stages: 1) initiation, 2) elongation, and 3) termination.
I. Initiation
The initiation of translation involves the binding and properly aligning the mRNA transcript to begin translation, aided by the action of initiation factors (IF).
The ribosomes in prokaryotes and eukaryotes both recognize distinct sequences between their mRNA transcripts.
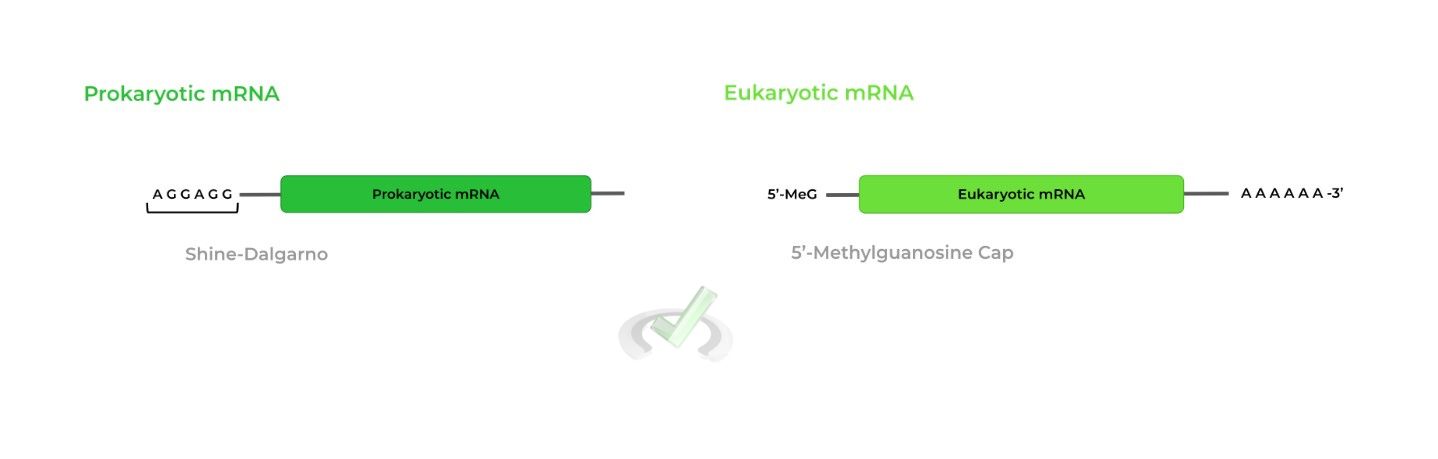
Prokaryotic ribosomes recognize the Shine-Dalgarno sequence in prokaryotic mRNA while eukaryotic ribosomes recognize the 5’-methylguanosine cap in eukaryotic mRNA!
II. Elongation
It’s best to visualize translation while the process is still ongoing as shown below! The left visual shows a newly arrived charged tRNA in the A site and a tRNA with a growing polypeptide in the P site.
An enzyme called peptidyl transferase will transfer the growing polypeptide chain to the charged tRNA located on the A site in order to attach the amino acid to the growing polypeptide.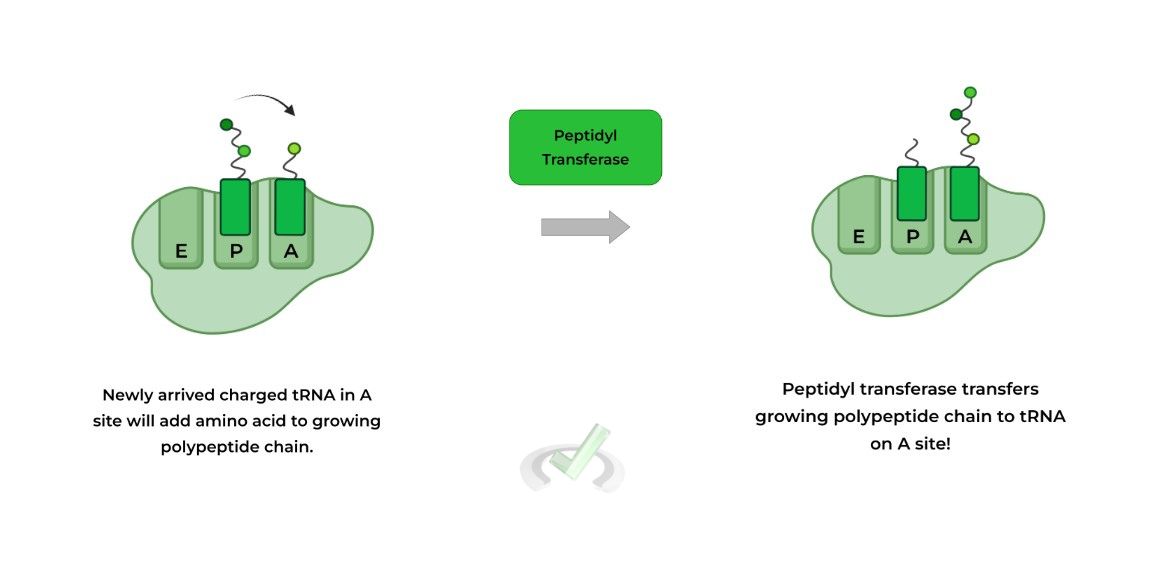
The then 2 tRNA molecules shift to the “left” which results in the uncharged tRNA – i.e. no amino acid attached – being placed in the E site and the tRNA with the growing polypeptide on the A site returns back to the P site.
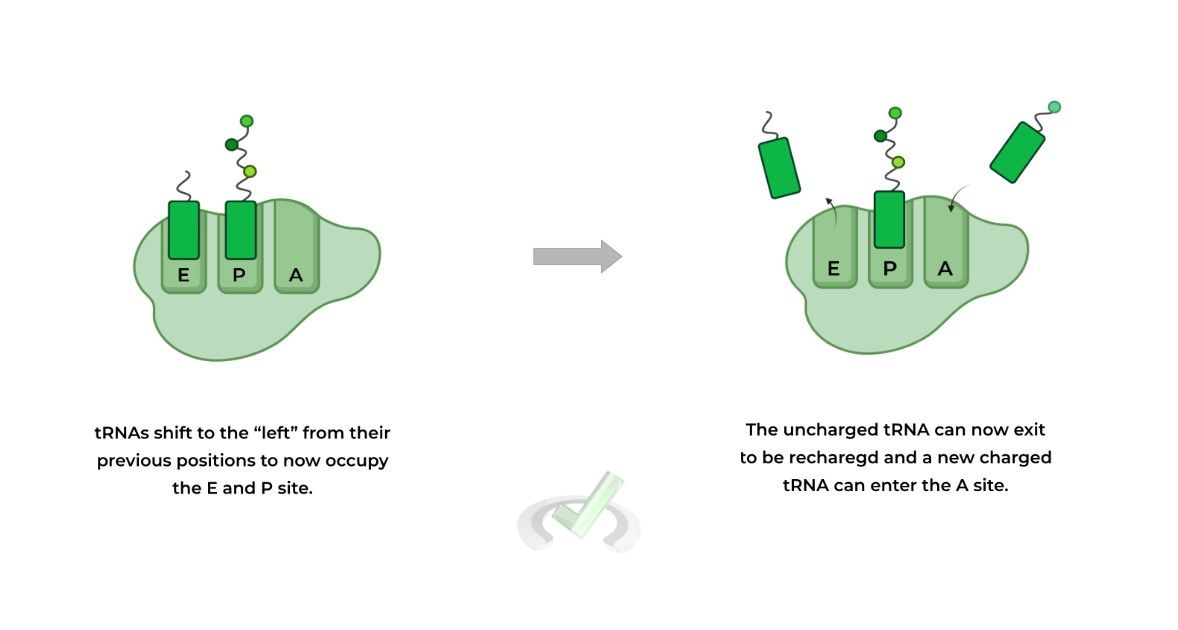
Now the uncharged tRNA in the E site can be released to be recharged with another amino acid and the process can begin again until terminated
III. Termination
Translation terminates upon the recognition of one of the stop codons: UAA, UAG, or UGA. This results in the recruitment of release and termination factors which aid in the hydrolysis and releasing of the polypeptide chain!
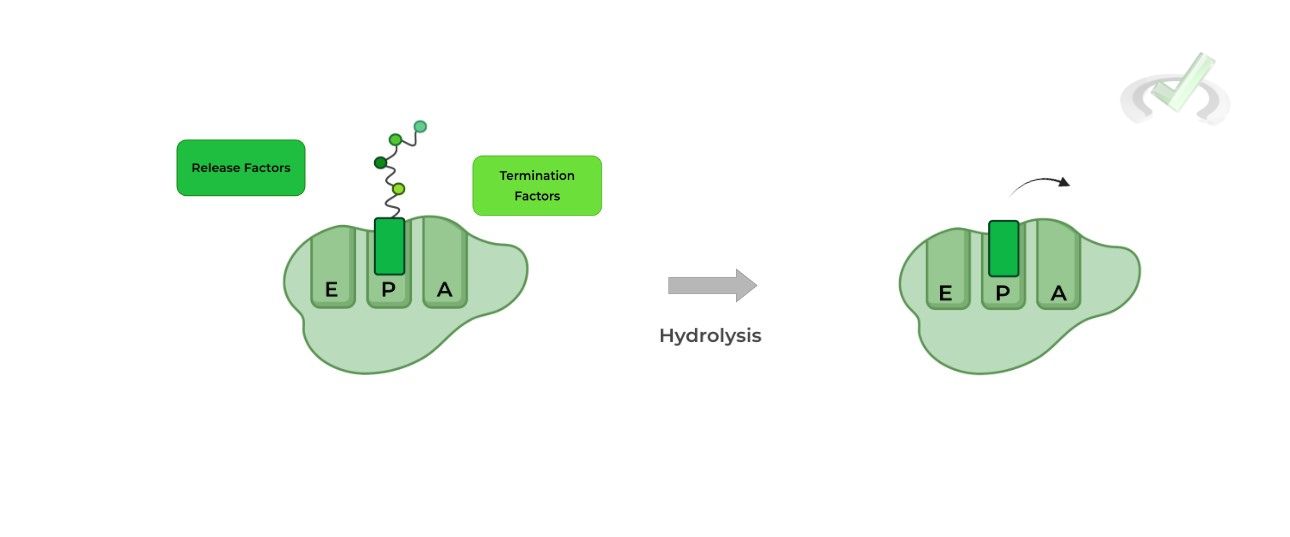
III. Bridge/Overlap
Similar to RNA transcription, there are also several post-translational modifications that proteins can undergo! Though there are many, we’ll primarily focus on 1) phosphorylation/dephosphorylation, 2) proteolysis, and 3) ubiquitination.
I. Post-Translational Modification: Phosphorylation/Dephosphorylation
This is probably the most common modification you’ve encountered studying other content, especially signaling pathways!
This usually involves a kinase or phosphatase which can phosphorylate or dephosphorylate serine, threonine, and tyrosine residues, respectively.
Note the amino acid residues! They are all viable targets for phosphorylation because of the hydroxyl group present in their side chains!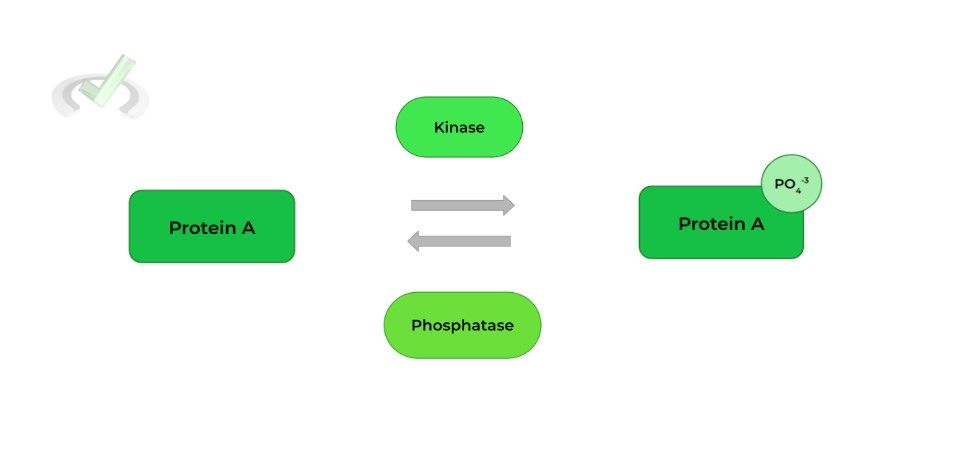
As a general rule of thumb, phosphorylation usually leads to protein ACTIVATION and dephosphorylation leads to protein DEACTIVATION (but there are some exceptions!).
II. Post-Translational Modification: Proteolysis
Oftentimes, the protein that results from translation still requires some further proteolytic cleavage in order to become activated and mature! This is actually used as a regulation mechanism in order to ensure that the protein only functions when needed too!
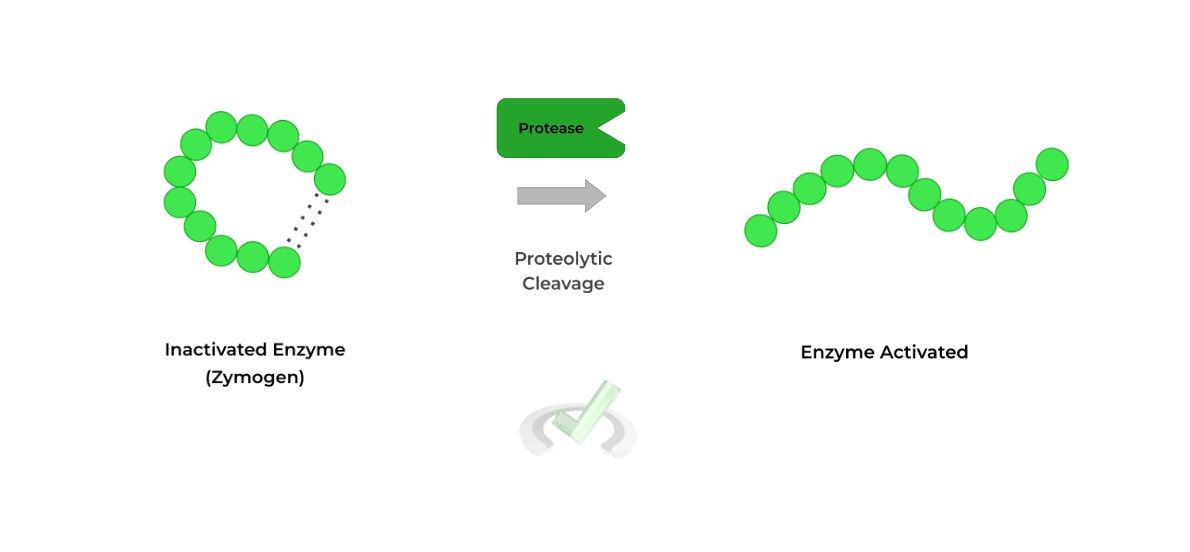
Some great examples of proteins that utilize these modifications are the digestive enzymes, such as pepsinogen and trypsinogen (zymogens), which are only cleaved and activated when food is present within the digestive tract.
III. Post-Translational Modification: Ubiquitination
In this proteinogenic modification, a ubiquitin group is covalently attached to the protein, targeting the protein for degradation by the proteasome.
This may seem counterintuitive because we want the protein to function. However, this is also used as a regulation mechanism in the case where the protein is no longer required to be present or the protein had defects during translation (i.e. wrong amino acids).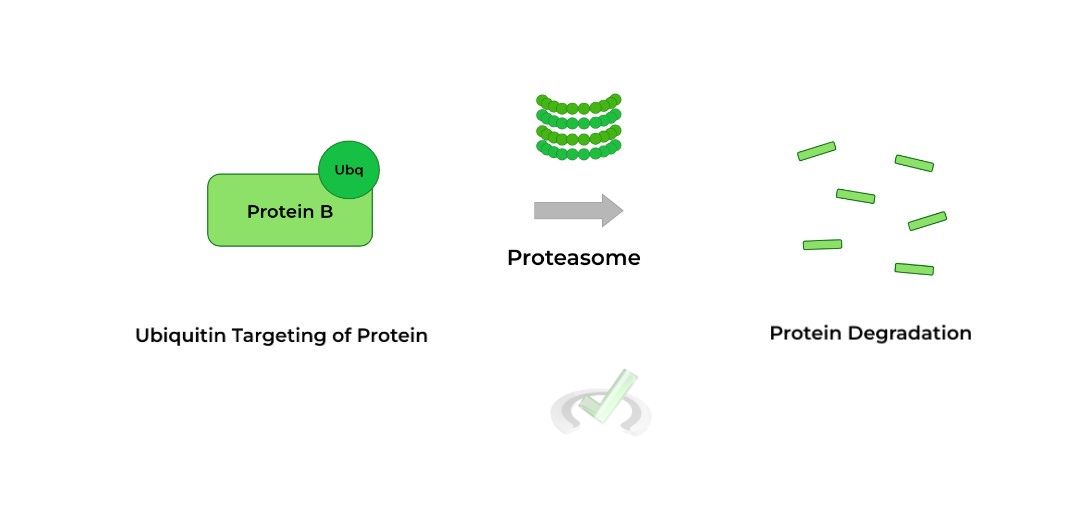
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Review of Ribosome Structure
Translation in the ribosome is governed by the various ribosomal sites which include the A (aminoacyl) site, P (peptide/petidyl) site, and the E (exit) site.
B. Process of Translation
The entire process of translation can actually be subdivided into 3 main stages: 1) initiation, 2) elongation, and 3) termination.
I. Initiation
Proteins called initiation factors (IF) will aid in the binding and aligning of the mRNA transcript to the ribosome.
Prokaryotic ribosomes recognize the Shine-Dalgarno sequence to facilitate binding while eukaryotic ribosomes recognize the 5’-methylguanosine cap.II. Elongation
When a new, charged tRNA enters into the A site, the enzyme peptidyl transferase transfers the growing polypeptide chain onto the new amino acid in the A site. Now, the tRNA in the P site is uncharged and the tRNA in the A site contains the polypeptide chain.
From here, the tRNAs “shift positions” to where the uncharged tRNA now lies in the E site for exit and the polypeptide tRNA is repositioned in the P site.
Now, another charged tRNA can enter into the A site, restarting the process until translation is terminated.
III. Termination
Upon recognition of one of the stop codons, UAA, UAG, or UGA, translation will terminate. Release and termination factors will then aid in the hydrolysis and release of the polypeptide chain.
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
After peptidyl transferase performs its function during translation, how many tRNAs are NOW present at this point of translation and how many have amino acid(s) attached to them?
A. 3; 2
B. 2; 2
C. 1; 1
D. 2; 1
Ans. D
Peptidyl transferase’s role is to transfer the growing polypeptide chain from the tRNA on the P site to the amino acid located on the A site, thus growing the chain. At this point, there are still 2 tRNAs present within the ribosome (one in the P site and one in the A site).
Likewise, the P site tRNA is now uncharged while the A site tRNA has the polypeptide chain. Now the tRNAs shift positions so that the uncharged tRNA is in the E site to exit and the polypeptide tRNA is now placed on the P site to recontinue the process.
Sample Practice Question 2
Part of the pathology of Alzheimer's Disease is the buildup of beta-amyloid protein plaques which should otherwise be eliminated as there is too much of this protein located between neurons. Which type of protein modification is most likely dysfunctional?
A. Ubiquitination
B. Phosphorylation
C. Dephosphorylation
D. Proteolysis
Ans. A
The targeting of a protein via ubiquitination is primarily used for the targeting of the protein for degradation via the proteasome. A default in this modification system is likely for Alzheimer’s as now the beta-amyloid protein plaques build up due to there not being a mode for degradation.
Phosphorylation/Dephosphorylation are mostly associated with signaling pathways and proteolysis, while cleaving proteins, is doing so for the purpose of further activation and maturation.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these