I. What is DNA Multiplication?
Though it’s usually reserved in the context of mitosis and producing daughter cells, the replication of certain DNA sequences and genes has many applications both in the research and real world, as we’ll give an example that affected everyone in the past years.
Though we were all struck and affected by the predicaments of the COVID-19 pandemic, it’s amazing just to think that some of these DNA duplication methods were used in the fight against this disease, from viral detection to life saving research to eventually producing a vaccine.
You’ll hopefully notice as well, luckily, that while covering these topics, there’ll be considerable overlap with the basic fundamentals of DNA replication and cell biology that maybe you’ve covered, just with a few alterations here and there!
II. Strategies for DNA Multiplication
Though there have been many approaches that have been developed, the 2 most well known and that we’ll be tested by the MCAT are 1) Polymerase Chain Reaction and 2) Gene Cloning. Let’s cover them in more detail!
A. Polymerase Chain Reaction
On face value, polymerase chain reactions are actually very similar to how DNA is replicated in the cell; in fact, it’s much simpler as you’ll see below! There are 3 main steps in PCR: 1) Denaturation. 2) Annealing, and 3) Elongation.
I. Denaturation
Given a dsDNA molecule, the first step is to denature the DNA double helix in order to expose the single DNA strands. In this case, heat is added in order to disrupt the hydrogen bonds between the nitrogenous bases.
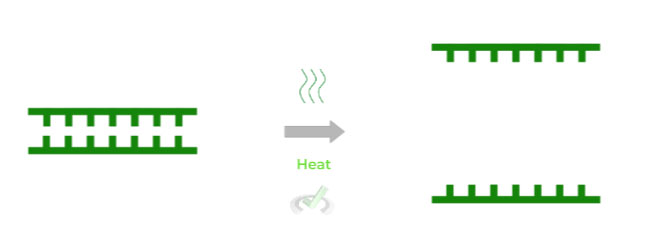
Notice how there’s not really a need to utilize DNA helicase because it’s much more efficient, both in time and resources, to use heat to denature the DNA molecule.
II. Annealing
Similar to DNA replication, DNA primers are included in order to initiate DNA strand synthesis. Recall that DNA polymerase cannot synthesize a new sequence without an already established sequence.
The process of the DNA primers binding to the single DNA strands is known as annealing and it’s now after this step that DNA elongation can take place!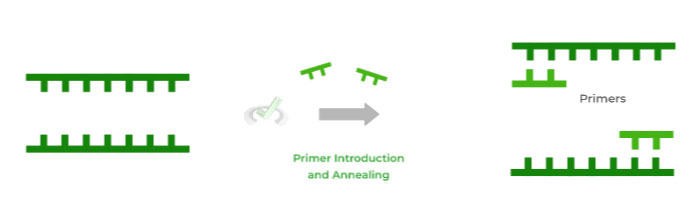
Note also that DNA primers are used, not the conventional RNA primers as in DNA replication. Now, unlike the RNA ones, these DNA primers don’t have to be removed!
III. Elongation
In this final step, elongation of the DNA strands can begin! Deoxyribonucleotides and a specialized DNA polymerase called Taq polymerase are added to the reaction to initiate DNA strand synthesis.
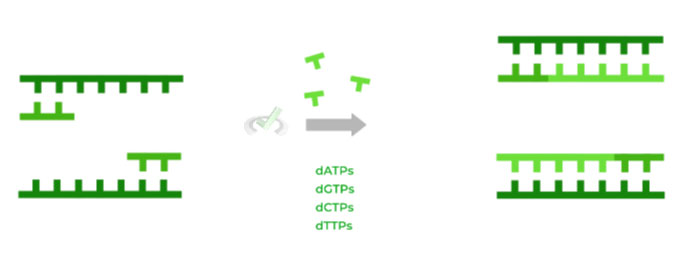
The normal DNA polymerases found in typical eukaryotes are not utilized in PCR because they have not evolved to withstand the high heat that’s required for PCR during denaturation.
As such, the Taq polymerase is used which has evolved to function in high temperatures, as it’s derived from the organism Thermus aquaticus, a bacteria that lives in the high temperature environment of the Yellowstone hot springs.
After many cycles of PCR, you’ll eventually end up with multiple DNA molecules, essentially doubling the DNA content after 1 round of PCR just like DNA replication! You can actually quantify the amount of DNA via the equation 2n, where n is the number of PCR cycles.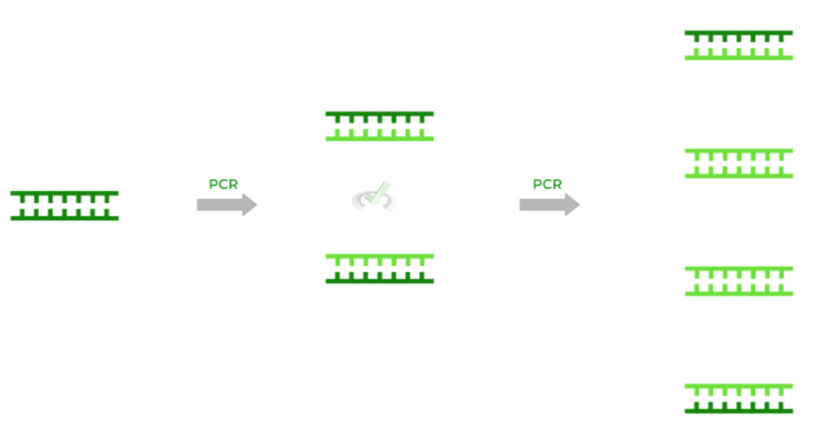
B. Recombinant Gene Cloning
This method is a bit more complicated, but has a wide variety of applications! Essentially, we’re using a bacterial organism in order to multiply our DNA gene of interest, which involves 1) Gene Insertion into Vector, 2) Vector Uptake into Bacteria, and 3) Bacterial Colony Growth.
I. Gene Insertion into Vector
The vector that’ll be used for gene cloning is a plasmid, which is essentially a small, circular DNA molecule. Through restriction enzymes, the gene can be inserted into the plasmid.
We’ll cover all the specific details about how restriction enzymes work in another article (i.e. palindromic sequences, sticky ends, etc.). For now, just know that the restriction enzymes work to aid in gene insertion!
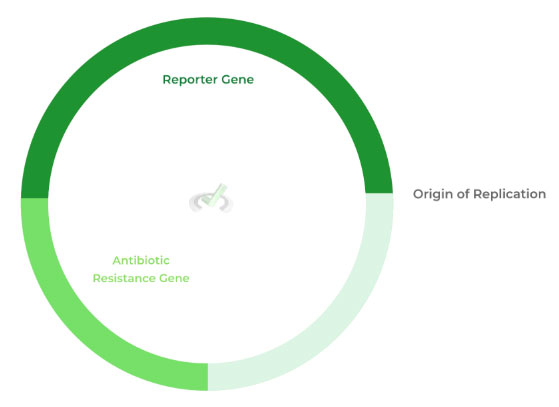
Let’s cover some of the basic features of the designed plasmid, as they’ll play important roles later on. The plasmid contains an origin of replication, an antibiotic resistance gene, and a reporter gene.
We’ll focus on the reporter gene for now as the origin of replication is self explanatory while reserving the antibiotic resistance gene for later.
The reporter gene usually codes for a fluorescent protein. An important aspect about the restriction enzymes is that they cut the reporter gene to where the gene can be inserted.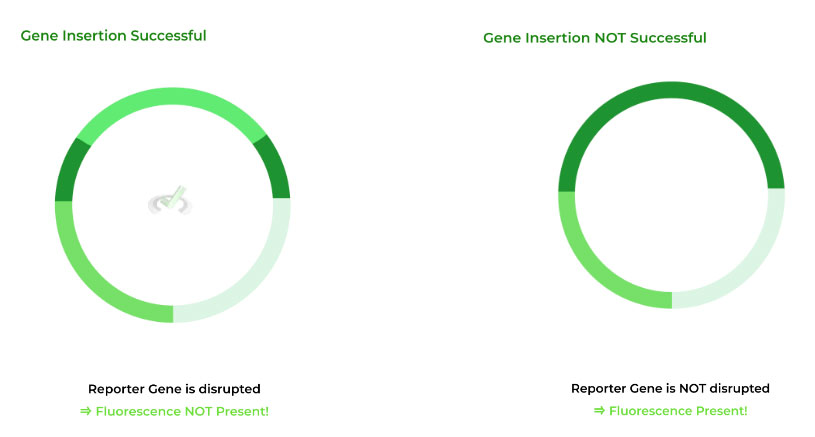
If the fluorescence is NOT PRESENT, the gene WAS inserted into the plasmid, because it was inserted into the reporter gene, disrupting the gene resulting in no expression of the fluorescence protein.
If the fluorescence is PRESENT, the gene WAS NOT inserted because the reported gene was not disrupted and thus could express the fluorescent protein!
II. Vector Uptake into Bacteria
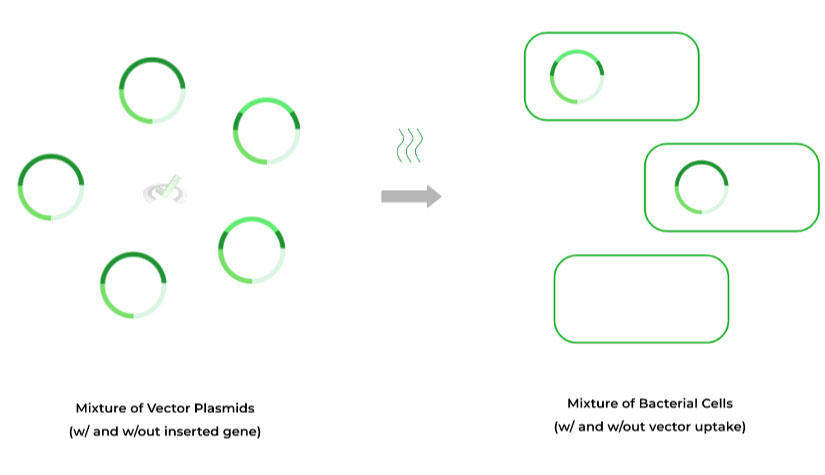
After the vector plasmid has had the gene of interest inserted (hopefully…), the next step is to transform the bacteria so that they can uptake our vector plasmids!
Some common ways to transform the bacteria are heat or electric shock (also called electroporation), which renders the bacterial membrane permeable for plasmid uptake.
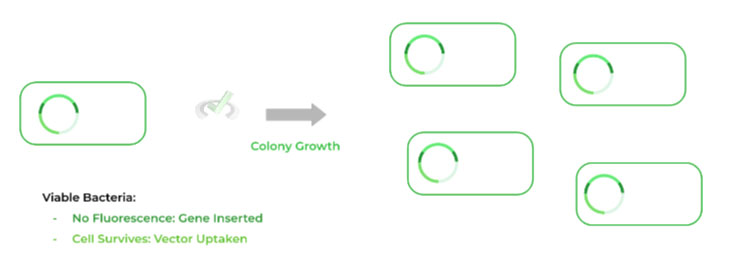
III. Bacterial Colony Growth
Before we can begin to grow the colonies and our DNA, we first need to perform the selection of the right bacteria. Here, we’re selecting for 2 things: 1) if the gene was inserted into plasmid and 2) if the plasmid was uptaken by the cell.
We already know to pick the bacteria with NO fluorescence, as this indicates that the gene was inserted because it disrupts the reporter gene.
Now, we can introduce an antibiotic to the bacteria to select for which cells up took the plasmids. Recall that the plasmids also carry an antibiotic resistance gene!
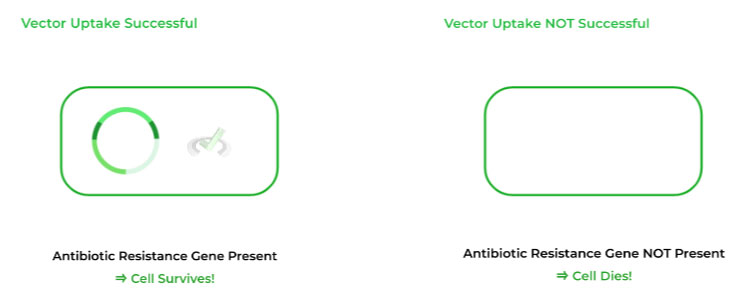
The cells that are still present after antibiotic introduction DID uptake the plasmid because they did contain the antibiotic resistance gene.
The cells that die after antibiotic introduction DID NOT uptake the plasmid because they did not have the antibiotic resistance gene.
From here, we choose the cells which are still present after antibiotic introduction and lack any fluorescence as these cells contain both the plasmid and inserted gene! The bacteria can now be grown and also multiply our inserted DNA!
III. Bridge/Overlap
We believe that one of the best ways to learn and review polymerase chain reactions are through relating the PCR steps to their DNA replication equivalents! Let’s take a look at them below!
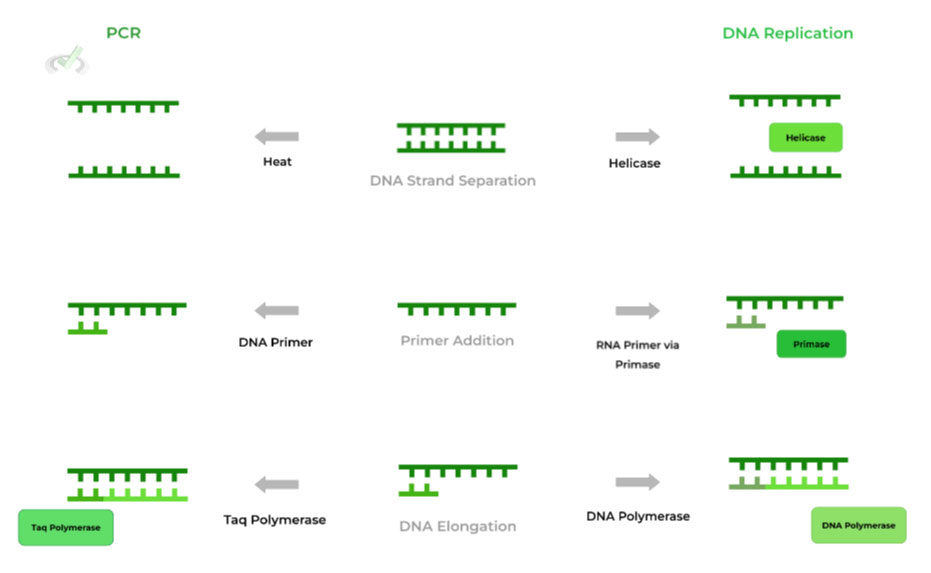
I. PCR Steps and DNA Replication Equivalents
As you’ve probably noticed, the 3 steps of denaturation, annealing, and elongation closely mimic the normal process of DNA replication in our cells! Below is a diagram that compares the 2 processes in terms of their outcomes!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Polymerase Chain Reaction
This technique of DNA multiplication is fairly similar in steps to DNA replication and includes 3 major steps: 1) Denaturation, 2) Annealing, and 3) Elongation.
I. Denaturation
The double stranded DNA must first be denatured to expose the single strands, which is accomplished by heating in order to disrupt the hydrogen bonds between the base pairs.
II. Annealing
After strand separation, DNA primers are added and anneal to the single strands in order to initiate elongation! Recall that DNA polymerase can only add nucleotides in an existing group of nucleotides!
III. Elongation
Because the primers have been added, DNA elongation can begin! In PCR, a special Taq polymerase is utilized as it’s evolved to withstand the high temperatures that PCR is performed under.
B. Gene Cloning
This technique is a bit more complicated as we’re using a bacterial organism to multiply our DNA, which involves 1) Gene Insertion into Vector, 2) Vector Uptake into Bacteria, and 3) Bacterial Colony Growth.
I. Gene Insertion into Vector
Via the action of restriction enzymes, our gene of interest is inserted into our vector plasmid! The vector plasmid has a specific design where it contains an origin of replication, an antibiotic resistance gene, and reporter gene.
The reporter gene usually codes for fluorescence; an important note is that the restriction enzymes cut the reporter gene so that our gene of interest can be inserted.
If fluorescence is NOT PRESENT, the gene WAS inserted into the plasmid because the gene insertion will disrupt the reporter gene and thus fluorescence expression.
If fluorescence is PRESENT, the gene WAS NOT inserted because the reporter gene was not disrupted allowing for fluorescence expression.II. Vector Uptake into Bacteria
The bacteria is then transformed in order to uptake the vector plasmid which contains our gene of interest (hopefully).
This is usually done via heat or electric shock to increase the permeability of the cell membrane to uptake the DNA.
III. Bacterial Colony Growth
Before the bacteria can be grown and replicate our DNA, we must first select for bacterial cells that up took the vector plasmid and have our gene of interest inserted.
We know to pick cells which show NO FLUORESCENCE, as these are the ones which have the gene inserted which disrupts the reporter gene. Now we can introduce an antibiotic to select for cells which up took the plasmid!
The cells present after antibiotic DID uptake the plasmid because they contain the antibiotic resistance gene.
The cells that died after antibiotic introduction DID NOT uptake the plasmid because they did not have the antibiotic resistance gene.
When we select for the viable bacterial cells, they can now be grown in colonies to multiply our gene of interest!
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
Which of the following would result in a bacterial cell showing fluorescence after antibiotic introduction?
A. Gene Insertion Successful; Vector Uptake Successful
B. Gene Insertion Successful; Vector Uptake Not Successful
C. Gene Insertion Not Successful; Vector Uptake Successful
D. Gene Insertion Not Successful; Vector Uptake Not Successful
Ans. C
We can infer that because fluorescence is shown that gene insertion WAS NOT successful because the fluorescent reporter gene was not disrupted and could be expressed.
Because fluorescence was shown AFTER antibiotic introduction, the cell survived indicating that vector uptake was successful because the cell contained the antibiotic resistance gene.
Sample Practice Question 2
Suppose a student was trying to multiply a gene sequence via gene cloning. However, he forgets to transform the bacteria with heat or electric shock but still introduces the antibiotic. Which of the following is expected to occur?
A. All the bacteria will show fluorescence.
B. Only some bacteria will show fluorescence.
C. All the bacteria will survive, but won’t have the vector plasmid.
D. All the bacteria will die.
Ans. D
Because the student forgot to transform the bacteria, the vector plasmids will not be uptaken by the cell. When the antibiotic is introduced, all the bacteria will die because they do not have the antibiotic resistance gene.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these