I. What are Restriction Enzymes?
Sometimes, especially in the sense of cell physiology, we generally view the cutting of nucleic acids as a bad thing! Think of all the havoc and problems we’d have if genomic DNA and messenger RNA were constantly cut and degraded!
However, we sometimes in fact do need to cut these nucleic acids for beneficial purposes of the cell as well! As a matter of fact, the repair mechanisms that act to repair DNA mutations utilize these cutting mechanisms in order to repair our damaged DNA!
One type of enzyme that contains these cutting mechanisms are restriction enzymes! These enzymes are crucial in many regards to cell function, but have high application in the field of biotechnology. We’ve actually covered one of their biotechnological uses which we’ll explore later!
II. Restriction Enzymes
Before actually getting into the actual function of restriction enzymes, let’s first look at some basic definitions and terms to get a better foundation!
A. Fundamental Terms for Restriction Enzymes
2 main terms and definitions to note about restriction enzymes and their function are 1) Endonuclease and 2) Palindromic Sequences. Let’s quickly and simply define these terms!
I. Endonuclease
An endonuclease is a type of enzyme which can cut/digest nucleic acids into fragments. Actually, another term for restriction enzymes are restriction endonucleases!
The restriction enzymes work to cleave nucleic acids by cleaving the phosphodiester backbone, releasing the fragment.
Recall that endonucleases and exonucleases differ by where they cut the nucleic acid! Endonuclease cut WITHIN the nucleic acid chain while exonucleases cut on the ENDS.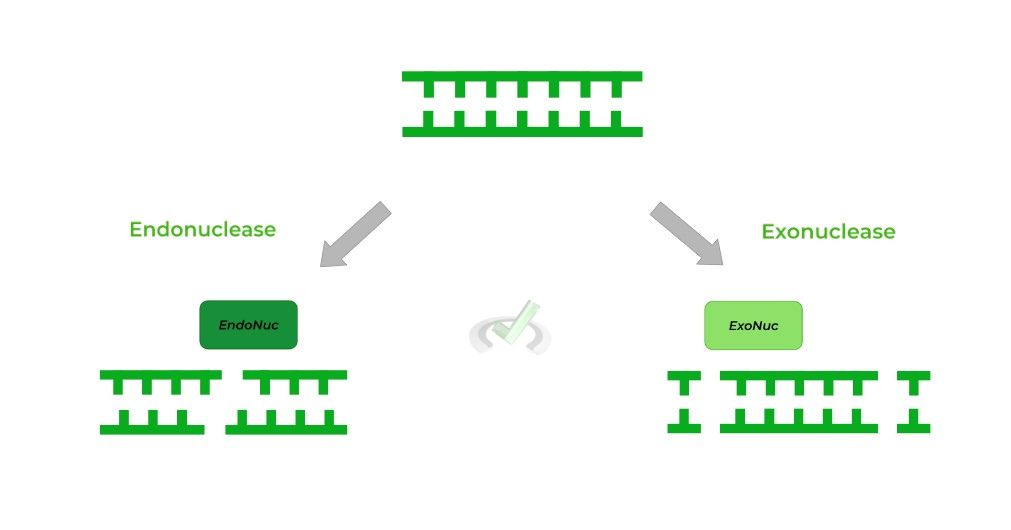
II. Palindromic Sequence
The benefits of restriction enzymes as opposed to other nucleases is their specificity! Whereas most nucleases cleave randomly, restriction enzymes only initiate their cuts at specific sequences called restriction sites or sequences.
Furthermore, these restriction sites have very specific motifs as they’re composed of palindromic sequences, which are when a 5’ to 3’ sequence on one strand is IDENTICAL to the 5’ to 3’ sequence on the complementary strand as shown below!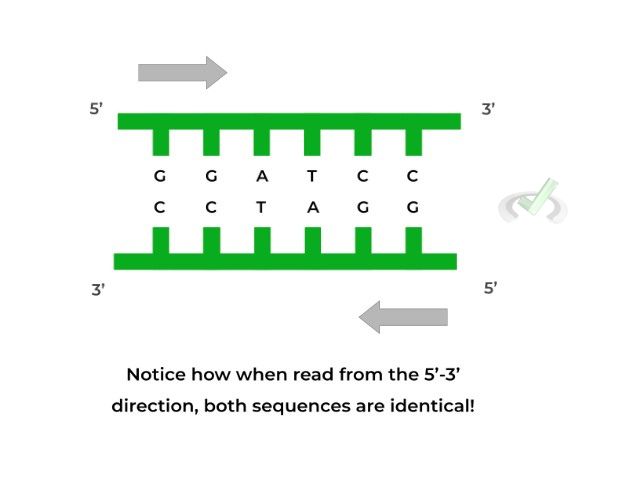
B. Function of Restriction Enzymes
As mentioned above, restriction enzymes cut at restriction sites which are composed of palindromic sequences! Each restriction enzyme has a specific palindromic sequence it recognizes and also cuts in a specific way!
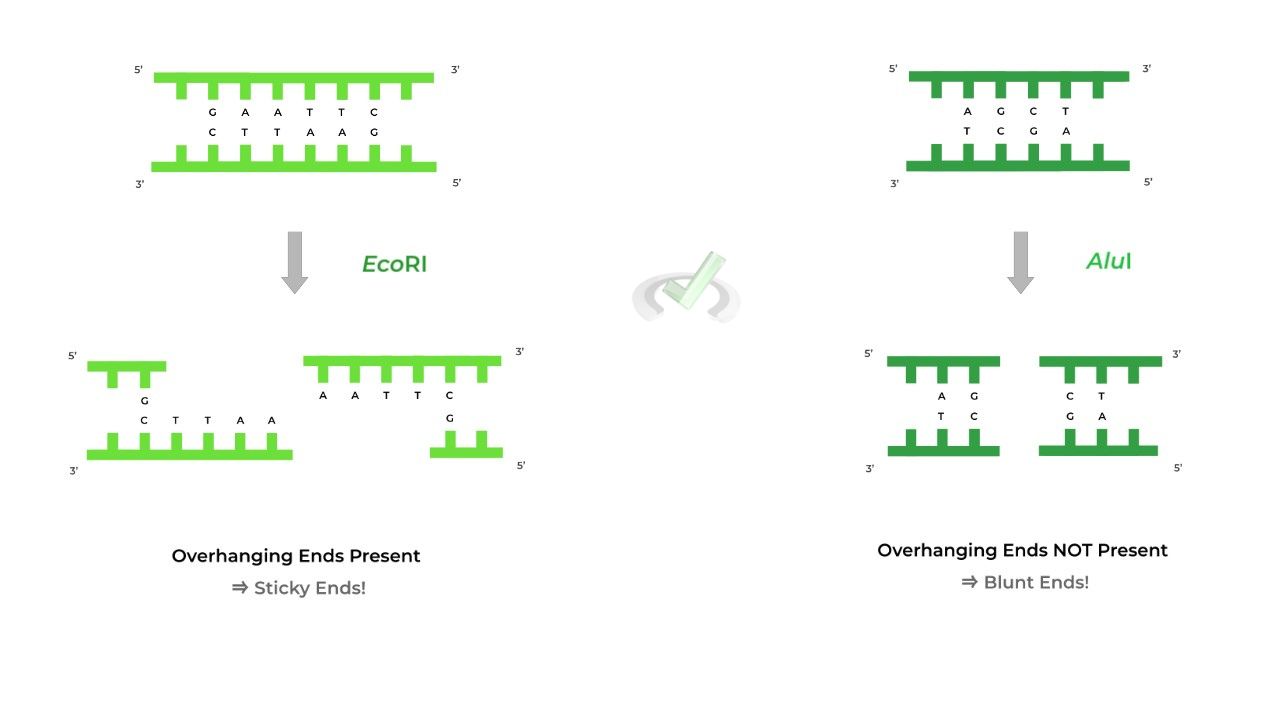
One result after restriction enzyme digestion are sticky ends (also called cohesive ends) which leave overhanging ends such as in the case of the EcoRI restriction enzyme!
Another resulting possibility is the presence of blunt ends, which does not leave overhanging ends, such as in the case of the AluI restriction enzyme.
C. Application for Gene Vectors
In another article covering strategies for DNA multiplication, we reviewed gene cloning and briefly mentioned the importance of restriction enzymes in the process. Let’s take an in depth look at how restriction enzymes aid in the process!
Recall that our gene of interest must be inserted into the vector plasmid. One special design of the plasmid is the inclusion of a palindromic recognition site where a restriction enzyme can cleave!
It’s important to choose a restriction enzyme and recognition site that will produce sticky ends, as it’ll be important when designing and inserting our gene.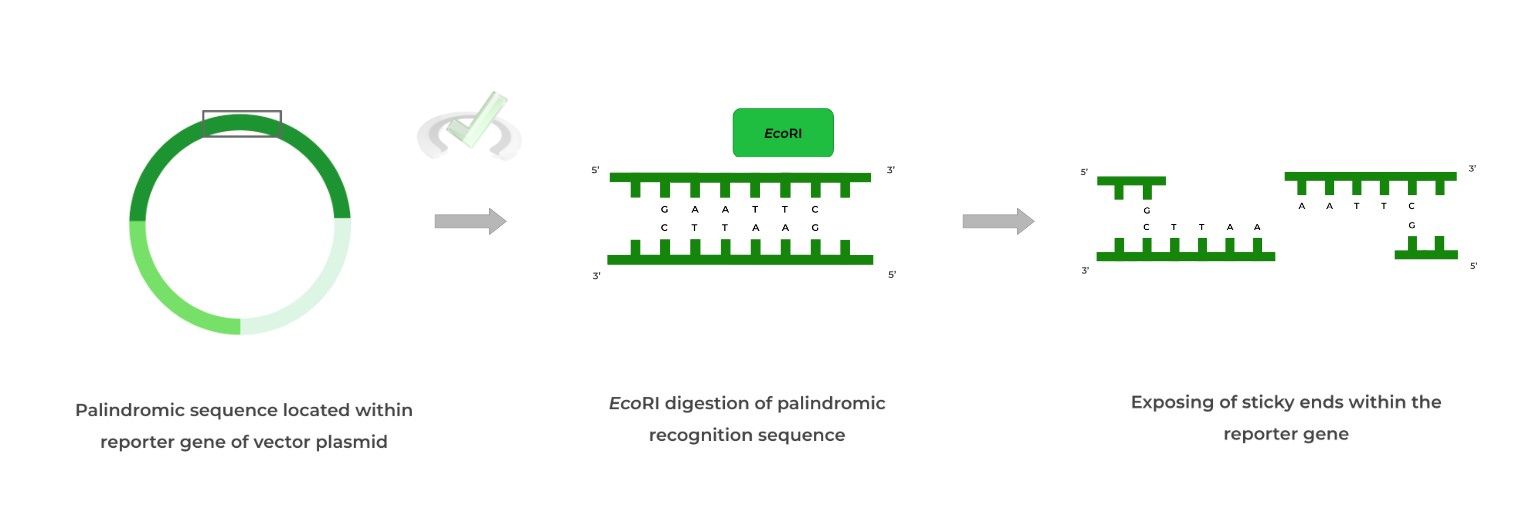
Now that the plasmid has been cleaved and the sticky ends are exposed, we can now insert our gene! Notice that in addition to the gene, we’ve added the complementary sticky end base pairs, which will anneal with the plasmid’s sticky ends, aiding in insertion!
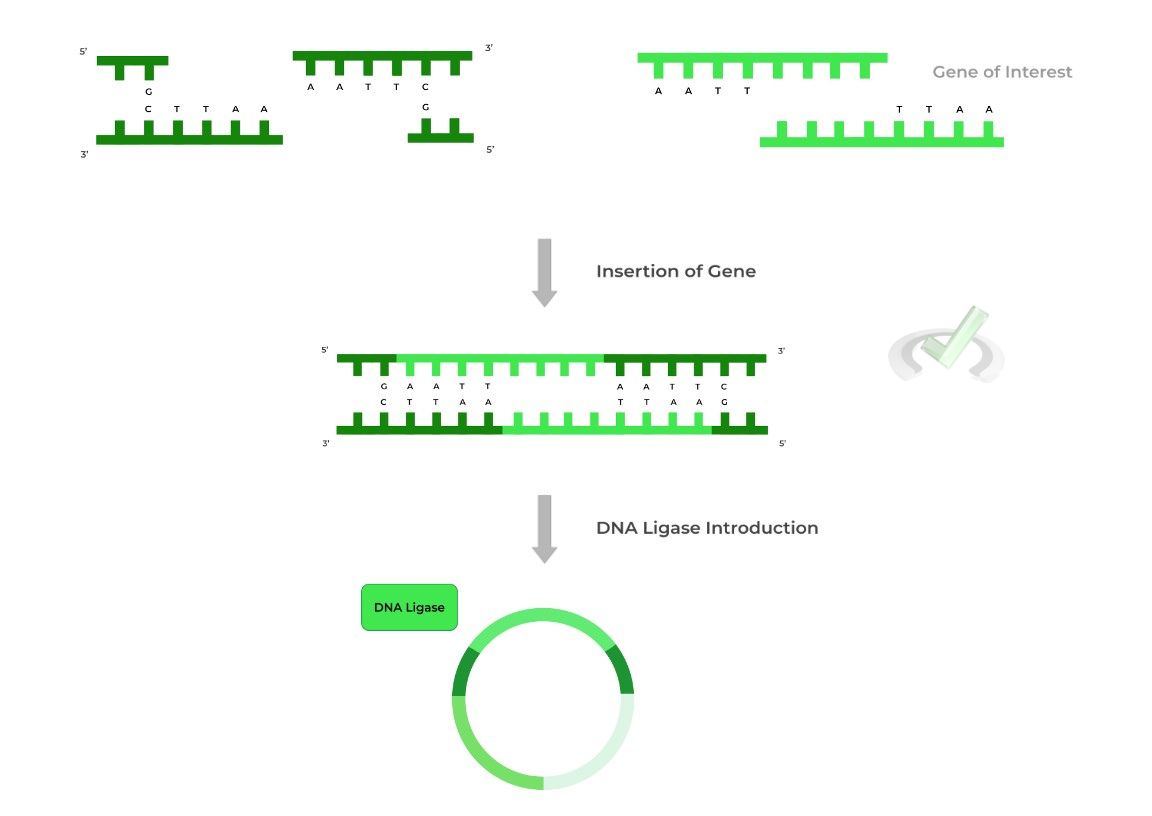
To finalize the insertion process, DNA ligase is added to ligate the inserted gene with the vector plasmid!
Note also that the palindromic recognition site must be within the reporter gene so that upon gene insertion, the reporter gene will be disrupted!
III. Bridge/Overlap
Similar to DNA synthesis, it may be helpful to highlight the mechanism for how restriction enzymes and other endonucleases/exonucleases cleave nucleic acids! Let’s review really quick!
I. Mechanism for Nuclease Catalyzed Hydrolysis
Nucleases are classified as hydrolases as they perform hydrolysis reactions resulting in the cleavage of the polyribonucleotide. If you’re familiar with the reaction mechanism of nucleotide addition, learning this will be easy!
Essentially, the H2O molecule will act as the nucleophile during the nucleophilic substitution and attack the phosphate group to release a free nucleotide.
However, the H20 molecule must first be deprotonated by a basic amino acid within the nuclease’s active site to be converted into a more nucleophilic hydroxyl group.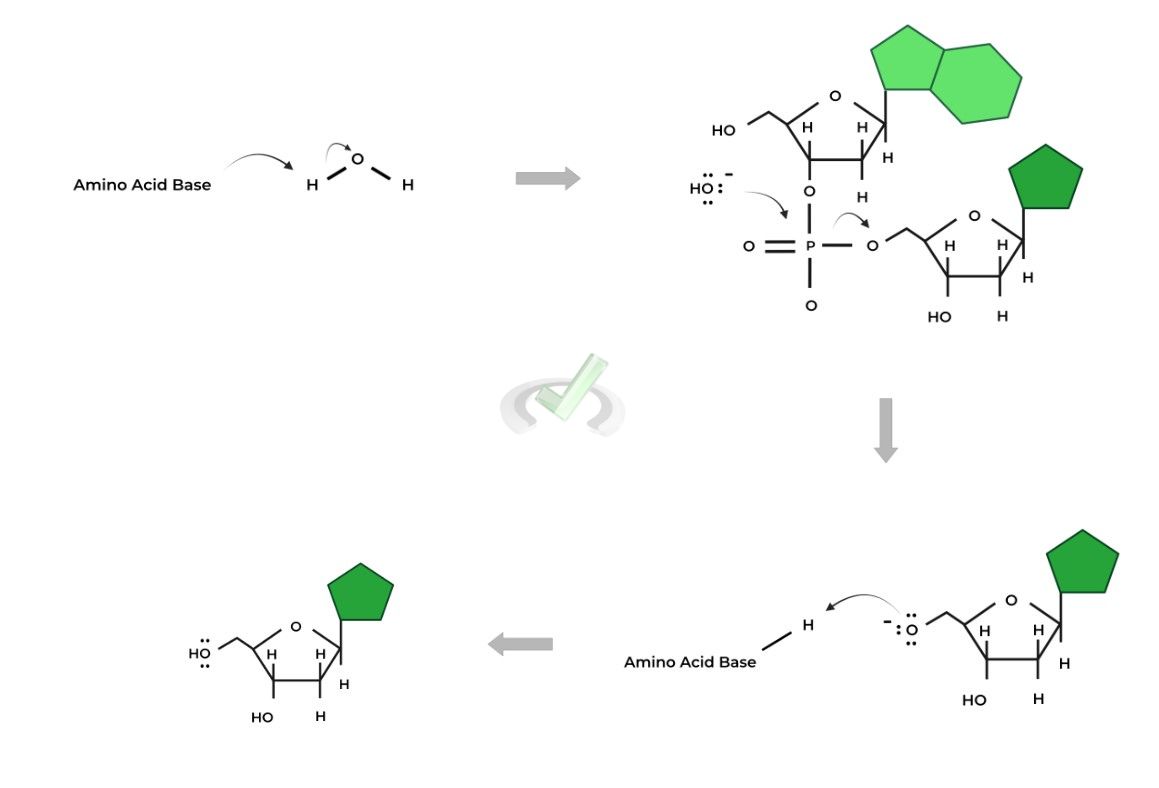
IV. Wrap Up/Overview
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. Fundamental Terms for Restriction Enzymes
When learning about restriction enzymes, there are 2 important terms and concepts which form the basis of their function: 1) Endonuclease and 2) Palindromic Sequences.
I. Endonuclease
Restriction enzymes are also called restriction endonucleases, as they work to cleave nucleic acids via hydrolysis. Note that endonucleases cut WITHIN the nucleic acids while exonucleases cut on the ENDS of nucleic acids.
II. Palindromic Sequences
Restriction enzymes will only cleave at specific sites called recognition sites, which are composed of palindromic sequences. Palindromic sequences occur when the 5’-3’ sequence on one strand is IDENTICAL to the 5’-3’ sequence on the complementary, antiparallel strand!
B. Function of Restriction Enzymes
There are 2 possible outcomes when restriction enzymes cut at the recognition sites. The cut DNA can either have overhanging strands termed sticky ends or no overhanging strands, termed blunt ends,
C. Application for Gene Vectors
In order for us to insert our gene into a vector plasmid, we must ensure that the palindromic, recognition site of a restriction enzyme is within the reporter gene.
The gene of interest also has sticky ends with the complementary base pairs which will bind to the sticky end base pairs of the reporter gene so that the gene of interest can be inserted!
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1:
Which of the following sequences will a restriction enzyme NOT recognize?
A. 5’ - AATT - 3’
3’ - TTAA - 5’
B. 5’ - ATCG - 3
3’ - TAGC - 5’
C. 5’ - CGTACG - 3’
3’ - GCATGC - 5’
D. 5’ - GATATC - 3’
3’ - CTATAG - 5’
Ans. B
Recall that restriction enzymes recognize palindromic sequences, which are DNA sequences where the 5’-3’ sequence on one strand is the same as the 5’-3’ sequence on the complementary strand!
Out of the choices above, choice B does not follow a palindromic sequence and will not be recognized by a restriction enzyme.
Sample Practice Question 2:
Given the following steps, which of the following list the correct sequence for gene insertion into a vector plasmid?
I. Recognition of Palindromic Sequence
II. DNA Ligase Action on Gene of Interest
III. Binding of Complementary Sticky Ends
A. I, III, II
B. I, II, III
C. II, III, I
D. III, II, I
Ans.A
In order for a gene of interest to be inserted, a restriction enzyme must first recognize and cut the palindromic sequence within the reporter gene. This results in the production of sticky ends and it’s at this point where the gene of interest can be inserted and have its complementary sticky ends bind.
Finally, the DNA ligase works to ligate the gene of interest into the vector plasmid via the synthesis of phosphodiester bonds.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these