I. What is Eukaryotic Gene Expression?
Welcome back again, assuming you’re coming from our “Gene Expression: Prokaryotes” article! Now that you’ve probably covered prokaryotic gene expression, let’s take a dive into the more complex gene expression of eukaryotes!
While definitely more complex, all these intricacies have a purpose! WIth a much higher variety of proteins, signaling pathways and, and cellular functions, the added complexity of eukaryotic gene expression is definitely warranted!
But don’t worry, we’re here to break it all down for you as simply and concisely as possible! You’ll probably notice that while going through the article, you’re also building off the foundations from what you’ve previously learned in gene structure and RNA transcription: let’s just now build off of them!
II. Control of Gene Expression in Eukaryotes
While there are various ways that gene expression can be regulated in eukaryotes, the 2 main ones to know for the MCAT are 1) transcription factors and 2) epigenetics. Let’s take a look at transcription factors first!
A. DNA Binding Proteins: Transcription Factors
Before talking about how transcription factors regulate gene expression, we first need a more detailed explanation of gene structure.
Neighboring the main gene sequence (the sequence that codes for the mRNA transcript), are cis-acting elements. Although initially a scary name, they simply refer to DNA sequences close to the gene that aid in transcriptional regulation.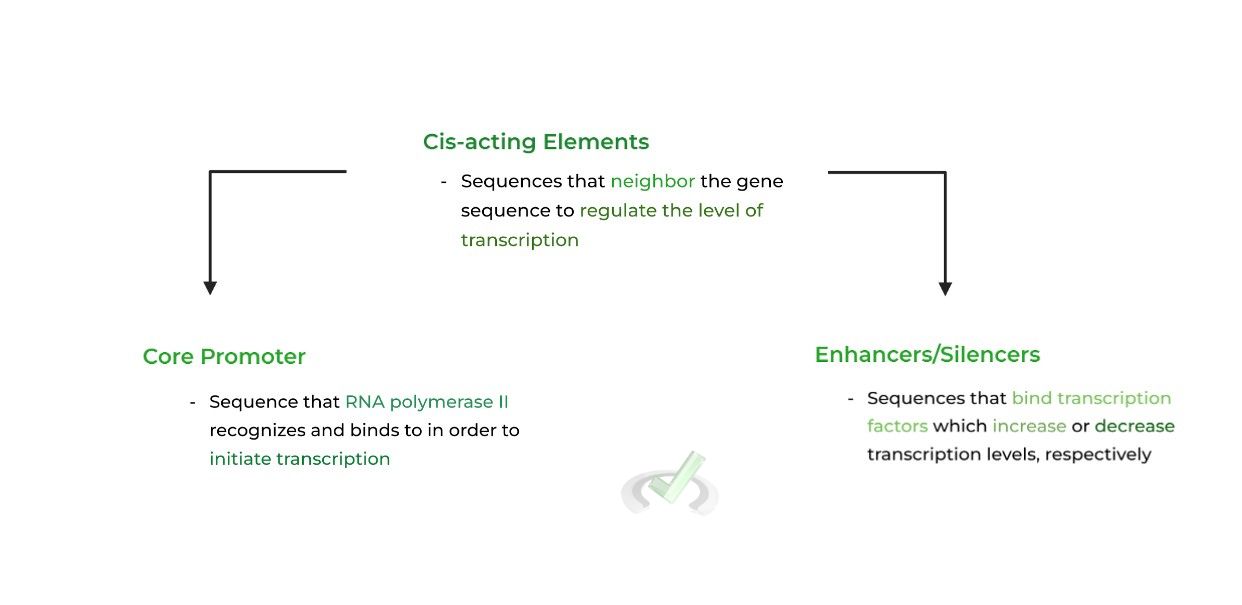
We’ve already discussed the TATA box as the main core promoter involved in eukaryotic gene expression in another article. Now, let’s focus on enhancers/silencers & transcription factors!
I. Enhancers and Silencers
As their name implies, enhancers and silencers promote and inhibit the transcription of a gene, usually located upstream of the gene sequence.
These sequences affect gene expression by serving as docking points, as they’ll bind to transcription factors, which are proteins that regulate the level of transcription by affecting the stability of RNA polymerase II.
Transcription “activating” factors will bind to enhancer regions, which promote transcription by stabilizing RNA polymerase II.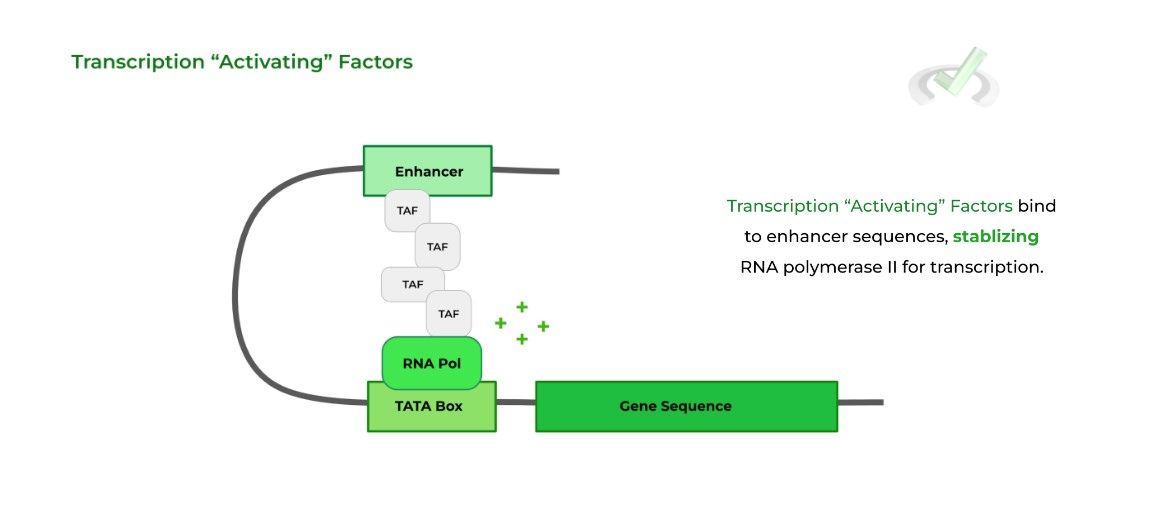
Likewise, transcription “inhibiting” factors bind to silencer regions, inhibiting transcription by destabilizing RNA polymerase II.
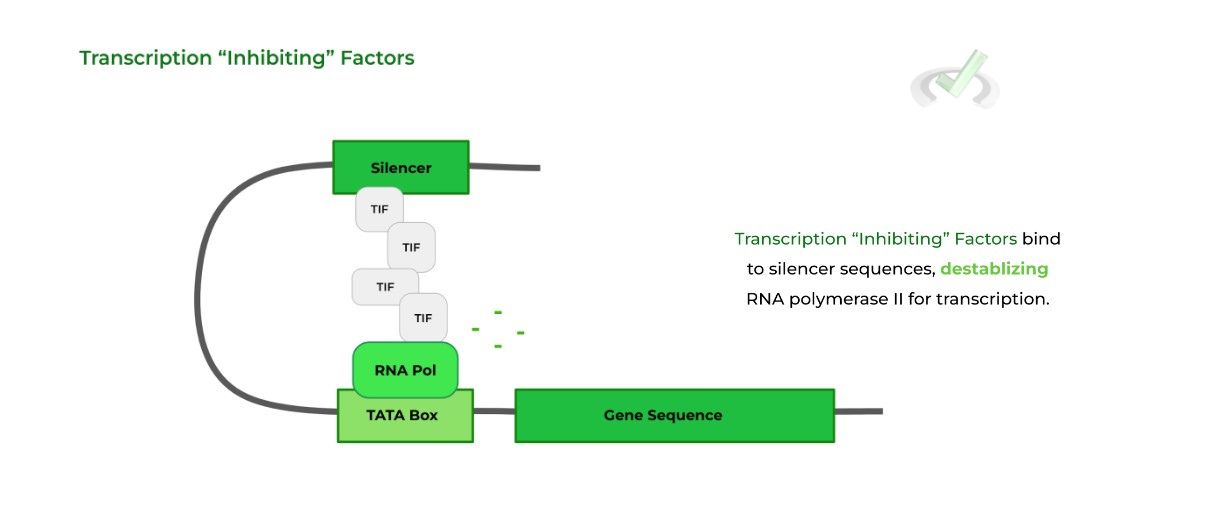
Note also how often the enhancers/silencers form a “DNA loop” to allow for the transcription factors to interact with the RNA polymerase.
Think of the enhancers/silencers like donors! Once the transcription factors bind to the sequences, they “donate” the proteins so that they can interact with the RNA polymerase and regulate transcription accordingly.B. Epigenetic Control of Gene Expression
Epigenetics deals with the regulation of gene expression specifically through chromatin remodeling! Recall that chromatin refers to the structure formed when DNA is wrapped around histone proteins.
Recall also that the basic structural units of chromatin are nucleosomes, which are composed of 4 pairs of histone proteins wrapped around proteins.
There are 2 main epigenetic modifications to chromatin that can alter whether it’s in a euchromatin (transcriptionally active) or heterochromatin state (transcriptionally inactive): 1) DNA methylation & 2) histone acetylation.I. DNA Methylation
This modification refers to the attachment of methyl groups to the DNA molecule. Enzymes called DNA methylases will attach methyl groups to cytosine bases of the gene sequence.
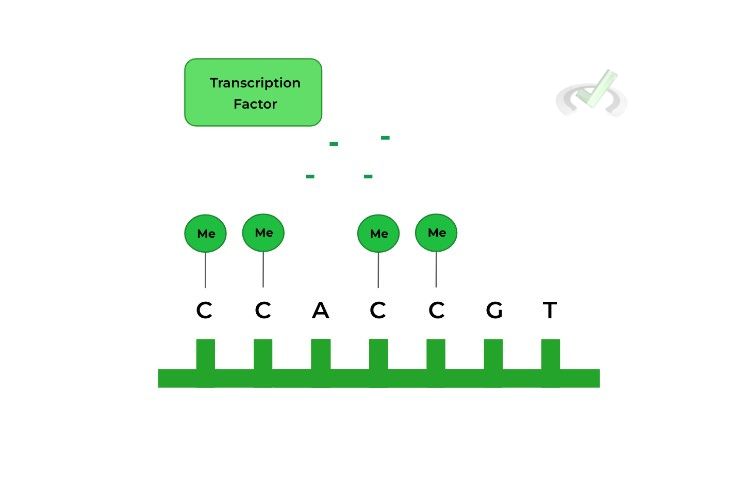
As shown above, the methylated cytosine bases on the DNA prevent the transcription factor interacting with the DNA, decreasing transcription! As a general rule of thumb, follow the relationships below:
DNA methylation results in a heterochromatin conformation (gene inactive) while DNA demethylation results in a euchromatin conformation (gene active!)II. Histone Acetylation
Similarly, this epigenetic modification involves the acetylation of histones, specifically at lysine residues via the enzyme acetylase.
Recall that lysine is positively charged! It turns out the acetylation of lysine DECREASES its positive charge, weakening the electrostatic interaction between the DNA & histone, “opening up” the DNA more.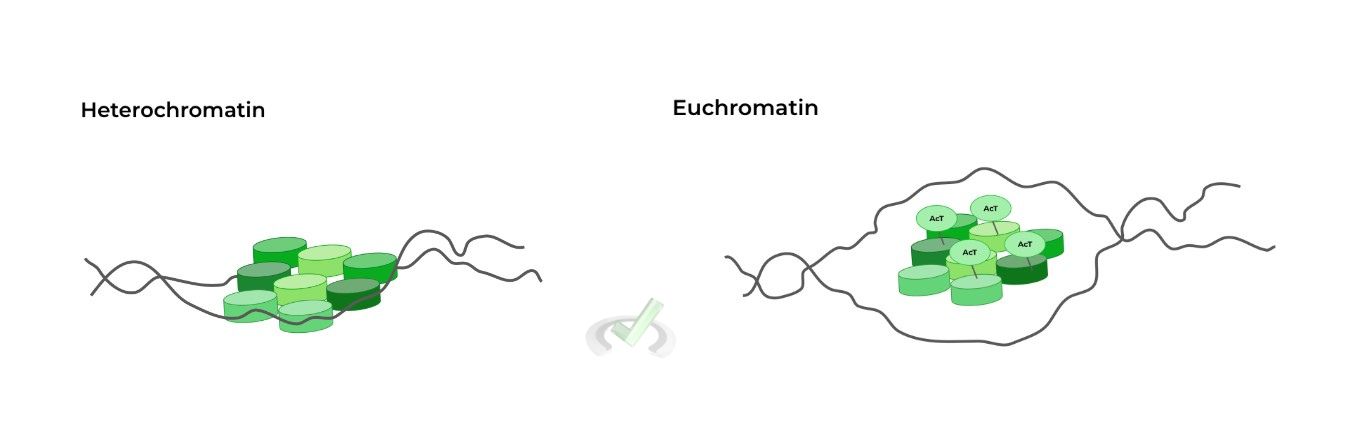
Because the chromatin now is in a euchromatin (“transcriptionally active”) state, transcription activating factors are now better suited to bind to the DNA and initiate transcription! Similarly, as a general rule of thumb, follow the relationships below:
Histone deacetylation results in a heterochromatin conformation (gene inactive) while histone acetylation results in a euchromatin conformation (gene active!)III. Bridge/Overlap
Transcription factors are heavily involved in signaling pathways, as they can be thought of as the bridge between the external signaling molecule (i.e. a hormone) and the internal expression of genes. Let’s take a look at the insulin-like growth factor 1 (IGF-1) molecule as an example.
I. Transcription Factors in Signaling Pathways
IGF-1, not surprisingly, is an external growth factor that can bind to cell receptors to initiate cell growth pathways. One of these pathways is actually the RAS-MAPK pathway, a well known growth pathway that’s often misregulated in cancer pathology.
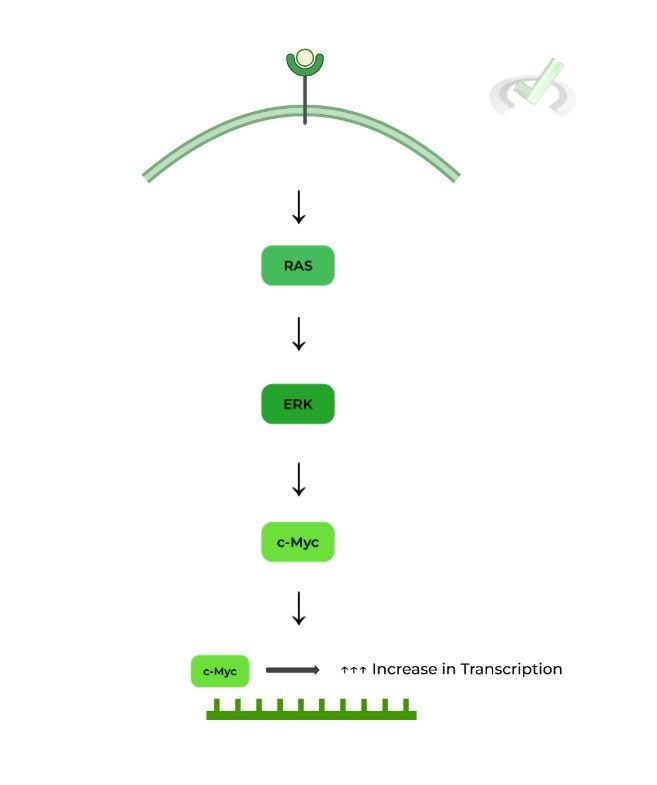
Here. the binding of IGF-1 to its respective receptor initiates in the following signal transduction pathway. This ultimately results in the activation, specifically to phosphorylation, of the c-Myc transcription factor, which binds to cell growth genes to increase transcription!
While there are some proteins missing, we wanted to simplify the pathway to more importantly show the role of transcription factors during signal transduction!
IV. Wrap Up/Key Terms
Let’s take this time to wrap up & concisely summarize what we covered above in the article!
A. DNA Binding Proteins: Transcription Factors
Transcription factors will bind to enhancers and silencers, which will then regulate transcription by affecting the stability of RNA polymerase II.
Transcription “activating” factors will bind to enhancer sequences and stabilize RNA polymerase II, increasing transcription levels.
Transcription “inhibiting” factors bind to silencer sequences and destabilize RNA polymerase II, decreasing transcription levels.B. Epigenetic Control of Gene Expression
This term refers to the regulation of gene expression, specifically through chromatin modification. The 2 main types of epigenetic modification are: 1) DNA methylation and 2) histone acetylation. In general, follow the rule of thumbs below:
I. DNA Methylation
DNA methylation results in a heterochromatin conformation (gene inactive) while DNA demethylation results in a euchromatin conformation (gene active!)
II. Histone Acetylation
Histone deacetylation results in a heterochromatin conformation (gene inactive) while histone acetylation results in a euchromatin conformation (gene active!)
V. Practice
Take a look at these practice questions to see and solidify your understanding!
Sample Practice Question 1
The interaction between histones and which component of DNA structure is affected upon histone acetylation?
A. Nitrogenous Base
B. Ribose Sugar
C. Phosphate Backbone
D. Deoxyribose Sugar
Ans. C
The acetylation of histone proteins at lysine residues causes the lysine to decrease in its positive charge. This results in a DECREASE in the electrostatic interaction between the positively charged lysine residues and the negatively charged DNA phosphate backbone.
Sample Practice Question 2
p53 is a gene that results in the overall INHIBITION of entrance into the cell cycle and thus cell proliferation. In the context of cancer, which wants to promote cell division, what epigenetic modifications of the p53 gene are most beneficial for the progression of cancer?
A. DNA Demethylation, Histone Acetylation
B. DNA Methylation, Histone Acetylation
C. DNA Demethylation, Histone Deacetylation
D. DNA Methylation, Histone Deacetylation
Ans. D
Because p53 is preventing the progression of cell proliferation and thus cancer, this gene needs to be INHIBITED in order for cancer to progress! The best combination of epigenetic modifications for p53 silencing are DNA Methylation and histone deacetylation, as now the chromatin is in a heterochromatin state.





 To help you achieve your goal MCAT score, we take turns hosting these
To help you achieve your goal MCAT score, we take turns hosting these